2W1W
 
 | | Native structure of a family 35 carbohydrate binding module from Clostridium thermocellum | | 分子名称: | CALCIUM ION, GLYCEROL, LIPOLYTIC ENZYME, ... | | 著者 | Gloster, T.M, Davies, G.J, Correia, M, Prates, J, Fontes, C, Gilbert, H.J. | | 登録日 | 2008-10-21 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W3J
 
 | | Structure of a family 35 carbohydrate binding module from an environmental isolate | | 分子名称: | CALCIUM ION, CARBOHYDRATE BINDING MODULE | | 著者 | Montainer, C, Flint, J, Gloster, T.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | 登録日 | 2008-11-12 | | 公開日 | 2009-01-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7RR0
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 222 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 222 Fab Heavy Chain, PDI 222 Fab Light Chain, ... | | 著者 | Pymm, P, Glukhova, A, Black, K.A, Tham, W.H. | | 登録日 | 2021-08-08 | | 公開日 | 2021-10-06 | | 最終更新日 | 2021-10-27 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
2VQP
 
 | | Structure of the matrix protein from human Respiratory Syncytial Virus | | 分子名称: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Money, V.A, McPhee, H.K, Sanderson, J.M, Yeo, R.P. | | 登録日 | 2008-03-18 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Surface Features of a Mononegavirales Matrix Protein Indicate Sites of Membrane Interaction.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4YQO
 
 | |
2VZ0
 
 | | Pteridine Reductase 1 (PTR1) from Trypanosoma Brucei in complex with NADP and DDD00066641 | | 分子名称: | 6-(4-methylphenyl)quinazoline-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE | | 著者 | Robinson, D.A, Thompson, S, Sienkiewicz, N, Fairlamb, A.H. | | 登録日 | 2008-07-29 | | 公開日 | 2009-09-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Development and Validation of a Cytochrome C Coupled Assay for Pteridine Reductase 1 and Dihydrofolate Reductase.
Anal.Biochem., 396, 2010
|
|
2VZQ
 
 | |
7RDS
 
 | | Structure of human NTHL1 | | 分子名称: | IRON/SULFUR CLUSTER, Isoform 3 of Endonuclease III-like protein 1 | | 著者 | Carroll, B.L, Zahn, K.E, Doublie, S. | | 登録日 | 2021-07-11 | | 公開日 | 2021-12-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Caught in motion: human NTHL1 undergoes interdomain rearrangement necessary for catalysis.
Nucleic Acids Res., 49, 2021
|
|
4WR8
 
 | | Macrophage Migration Inhibitory Factor in complex with a biaryltriazole inhibitor (3b-180) | | 分子名称: | 4-[4-(quinolin-2-yl)-1H-1,2,3-triazol-1-yl]phenol, Macrophage migration inhibitory factor, SODIUM ION, ... | | 著者 | Robertson, M.J, Baxter, R.H.G, Jorgensen, W.L. | | 登録日 | 2014-10-23 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Design, synthesis, and protein crystallography of biaryltriazoles as potent tautomerase inhibitors of macrophage migration inhibitory factor.
J.Am.Chem.Soc., 137, 2015
|
|
7RDT
 
 | | Structure of human NTHL1 - linker 1 chimera | | 分子名称: | IRON/SULFUR CLUSTER, Isoform 3 of Endonuclease III-like protein 1 | | 著者 | Carroll, B.L, Doublie, S. | | 登録日 | 2021-07-11 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Caught in motion: human NTHL1 undergoes interdomain rearrangement necessary for catalysis.
Nucleic Acids Res., 49, 2021
|
|
2WYK
 
 | | SiaP in complex with Neu5Gc | | 分子名称: | N-glycolyl-beta-neuraminic acid, SIALIC ACID-BINDING PERIPLASMIC PROTEIN SIAP, THIOCYANATE ION | | 著者 | Fischer, M, Hubbard, R.E. | | 登録日 | 2009-11-16 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Water networks can determine the affinity of ligand binding to proteins.
J.Am.Chem.Soc., 2019
|
|
4X1M
 
 | |
3G7M
 
 | | Structure of the thaumatin-like xylanase inhibitor TLXI | | 分子名称: | GLYCEROL, SODIUM ION, Xylanase inhibitor TL-XI | | 著者 | Vandermarliere, E, Courtin, C.M, Lammens, W, Schoepe, J, Strelkov, S.V. | | 登録日 | 2009-02-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Crystal structure of the noncompetitive xylanase inhibitor TLXI, member of the small thaumatin-like protein family.
Proteins, 78, 2010
|
|
4X0K
 
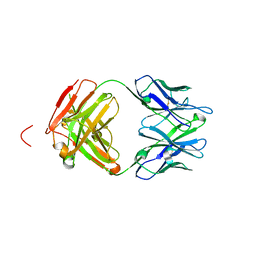 | | Engineered Fab fragment specific for EYMPME (EE) peptide | | 分子名称: | Fab fragment heavy chain, Fab fragment light chain | | 著者 | Johnson, J.L, Lieberman, R.L. | | 登録日 | 2014-11-21 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structural and biophysical characterization of an epitope-specific engineered Fab fragment and complexation with membrane proteins: implications for co-crystallization.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7R9C
 
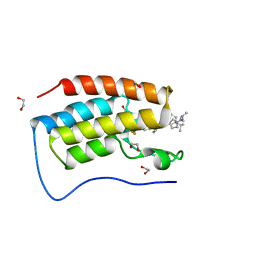 | | Cocrystal of BRD4(D1) with N,N-dimethyl-2-[(3R)-3-(5-{2-[2-methyl-5-(propan-2-yl)phenoxy]pyrimidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-1-yl)pyrrolidin-1-yl]ethan-1-amine | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | 著者 | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-06-29 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RPM
 
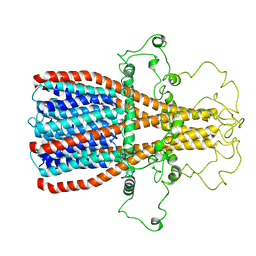 | |
4WN5
 
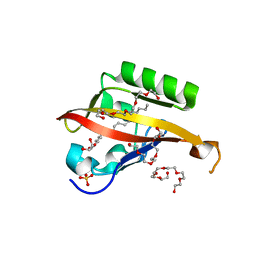 | | Crystal structure of the C-terminal Per-Arnt-Sim (PASb) of human HIF-3alpha9 bound to 18:1-1-monoacylglycerol | | 分子名称: | HEXAETHYLENE GLYCOL, Hypoxia-inducible factor 3-alpha, MONOVACCENIN, ... | | 著者 | Fala, A.M, Oliveira, J.F, Dias, S.M, Ambrosio, A.L. | | 登録日 | 2014-10-10 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Unsaturated fatty acids as high-affinity ligands of the C-terminal Per-ARNT-Sim domain from the Hypoxia-inducible factor 3 alpha.
Sci Rep, 5, 2015
|
|
4WPE
 
 | |
3GIP
 
 | | Crystal structure of N-acyl-D-Glutamate Deacylase from Bordetella Bronchiseptica complexed with zinc, acetate and formate ions. | | 分子名称: | ACETIC ACID, FORMIC ACID, N-acyl-D-glutamate deacylase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Almo, S.C. | | 登録日 | 2009-03-05 | | 公開日 | 2009-09-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Annotating enzymes of uncertain function: the deacylation of D-amino acids by members of the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
7RG8
 
 | | Crystal Structure of a Stable Heparanase Mutant | | 分子名称: | ACETATE ION, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit, ... | | 著者 | Whitefield, C, Hong, N.S, Jackson, C.J. | | 登録日 | 2021-07-14 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Computational design and experimental characterisation of a stable human heparanase variant.
Rsc Chem Biol, 3, 2022
|
|
2WIN
 
 | | C3 convertase (C3bBb) stabilized by SCIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, ... | | 著者 | Wu, J, Janssen, B.J, Gros, P. | | 登録日 | 2009-05-13 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor.
Nat. Immunol., 10, 2009
|
|
4X2I
 
 | | Discovery of benzotriazolo diazepines as orally-active inhibitors of BET bromodomains: Crystal structure of BRD4 with CPI-13 | | 分子名称: | (4R)-6-(4-chlorophenyl)-1,4-dimethyl-5,6-dihydro-4H-[1,2,4]triazolo[4,3-a][1,5]benzodiazepine, Bromodomain-containing protein 4, FORMIC ACID | | 著者 | Bellon, S.F, Jayaram, H, Poy, F. | | 登録日 | 2014-11-26 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Discovery of Benzotriazolo[4,3-d][1,4]diazepines as Orally Active Inhibitors of BET Bromodomains.
Acs Med.Chem.Lett., 7, 2016
|
|
7RD0
 
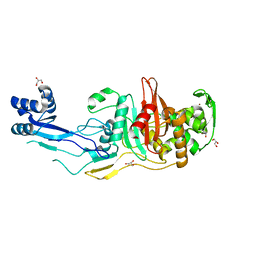 | |
3GD2
 
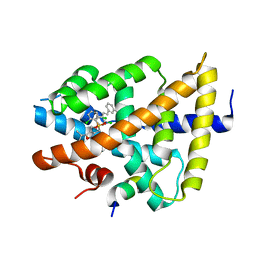 | | isoxazole ligand bound to farnesoid X receptor (FXR) | | 分子名称: | 3-[(E)-2-(2-chloro-4-{[3-{[(R)-(2,6-dichlorophenyl)(hydroxy)-lambda~4~-sulfanyl]methyl}-5-(1-methylethyl)isoxazol-4-yl]methoxy}phenyl)ethenyl]benzoic acid, Bile acid receptor, activator peptide | | 著者 | Madauss, K.P, Williams, S.P, Deaton, D.N, Wisely, G.B, Mcfadyen, R.B. | | 登録日 | 2009-02-23 | | 公開日 | 2009-07-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Substituted isoxazole analogs of farnesoid X receptor (FXR) agonist GW4064.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7RCW
 
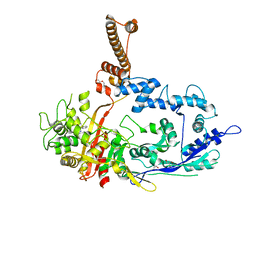 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2021-07-08 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
