6SCI
 
 | | Structure of AdhE form 1 | | 分子名称: | Aldehyde-alcohol dehydrogenase, FE (III) ION | | 著者 | Lovering, A.L, Bragginton, E. | | 登録日 | 2019-07-24 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | High-resolution structure of the alcohol dehydrogenase domain of the bifunctional bacterial enzyme AdhE.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7ZLE
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP | | 分子名称: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Roversi, P, Zitzmann, N, Bayo, Y, Le Cornu, J.D. | | 登録日 | 2022-04-14 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.823 Å) | | 主引用文献 | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein quality control checkpoint
To Be Published
|
|
7ZXW
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with the 5-[(morpholin-4-yl)methyl]quinolin-8-ol inhibitor | | 分子名称: | 5-(morpholin-4-ylmethyl)quinolin-8-ol, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | 著者 | Le Cornu, J.D, Ibba, R, Roversi, P, Zitzmann, N. | | 登録日 | 2022-05-23 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.246 Å) | | 主引用文献 | Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint.
Front Mol Biosci, 9, 2022
|
|
5CEL
 
 | | CBH1 (E212Q) CELLOTETRAOSE COMPLEX | | 分子名称: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | 著者 | Divne, C, Stahlberg, J, Jones, T.A. | | 登録日 | 1997-09-24 | | 公開日 | 1997-12-24 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
6I0I
 
 | |
4LWD
 
 | | Human CARMA1 CARD domain | | 分子名称: | Caspase recruitment domain-containing protein 11, MAGNESIUM ION, SULFATE ION | | 著者 | Zheng, C, Wu, H. | | 登録日 | 2013-07-26 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly.
Mol.Cell, 51, 2013
|
|
6TRF
 
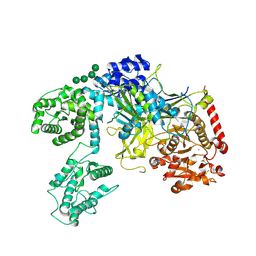 | | Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) purified from cells treated with kifunensine. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | 著者 | Roversi, P, Zitzmann, N. | | 登録日 | 2019-12-18 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (4.106 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TS2
 
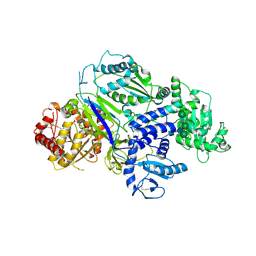 | | Truncated version of Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) lacking domain TRXL2 (417-650). | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | 著者 | Roversi, P, Zitzmann, N. | | 登録日 | 2019-12-19 | | 公開日 | 2020-01-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (5.74 Å) | | 主引用文献 | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6H41
 
 | |
7MTB
 
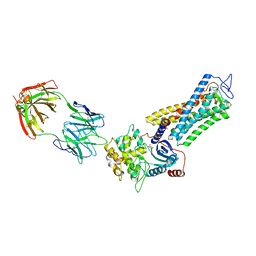 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab6 | | 分子名称: | Fab6 heavy chain, Fab6 light chain, RETINAL, ... | | 著者 | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | 登録日 | 2021-05-13 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT8
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin | | 分子名称: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | 著者 | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | 登録日 | 2021-05-13 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MTA
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab1 | | 分子名称: | Fab1 Heavy chain, Fab1 Light chain, RETINAL, ... | | 著者 | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | 登録日 | 2021-05-13 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
7MT9
 
 | | Rhodopsin kinase (GRK1) in complex with rhodopsin | | 分子名称: | RETINAL, Rhodopsin, Rhodopsin kinase GRK1, ... | | 著者 | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | 登録日 | 2021-05-13 | | 公開日 | 2021-07-07 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
4H9G
 
 | | Probing EF-Tu with a very small brominated fragment library identifies the CCA pocket | | 分子名称: | 5-bromofuran-2-carboxylic acid, AMMONIUM ION, Elongation factor Tu-A, ... | | 著者 | Groftehauge, M.K, Therkelsen, M, Taaning, R.H, Skrydstrup, T, Morth, J.P, Nissen, P. | | 登録日 | 2012-09-24 | | 公開日 | 2013-09-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6FSN
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-glucose (conformation 1) | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Roversi, P, Le Cornu, J.D, Hill, J, Alonzi, D.S, Zitzmann, N. | | 登録日 | 2018-02-19 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint.
Front Mol Biosci, 2022
|
|
6GL4
 
 | | Structure of GluA2o ligand-binding domain (S1S2J) in complex with glutamate and sodium bromide at 1.95 A resolution | | 分子名称: | ACETATE ION, BROMIDE ION, GLUTAMIC ACID, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-05-22 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.948 Å) | | 主引用文献 | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
3OK8
 
 | | I-BAR OF PinkBAR | | 分子名称: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 2, GLYCEROL | | 著者 | Boczkowska, M, Rebowski, G, Saarikangas, J, Lappalainen, P, Dominguez, R. | | 登録日 | 2010-08-24 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6GIV
 
 | | Structure of GluA2-N775S ligand-binding domain (S1S2J) in complex with glutamate and Rubidium Bromide at 1.75 A resolution | | 分子名称: | BROMIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2018-05-15 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
3TTZ
 
 | | Crystal structure of a topoisomerase ATPase inhibitor | | 分子名称: | 2-[(3S,4R)-4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}-3-fluoropiperidin-1-yl]-1,3-thiazole-5-carboxylic acid, DNA gyrase subunit B, MAGNESIUM ION | | 著者 | Boriack-Sjodin, P.A, Read, J, Eakin, A.E, Sherer, B.A. | | 登録日 | 2011-09-15 | | 公開日 | 2011-11-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Pyrrolamide DNA gyrase inhibitors: Optimization of antibacterial activity and efficacy.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8DAD
 
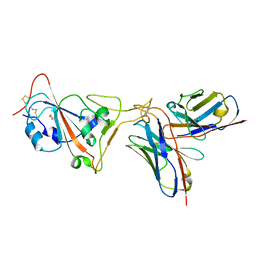 | | SARS-CoV-2 receptor binding domain in complex with AZ090 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AZ090 Fab Heavy Chain, AZ090 Fab Light Chain, ... | | 著者 | Zong, S, Wang, Z, Gaebler, C, Nussenzweig, M. | | 登録日 | 2022-06-13 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | SARS-CoV-2 receptor binding domain in complex with AZ090 Fab
To Be Published
|
|
4A56
 
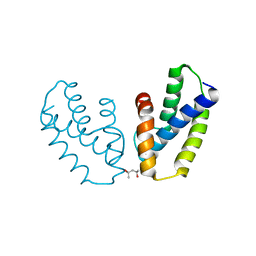 | | Crystal structure of the type 2 secretion system pilotin from Klebsiella Oxytoca | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, PULLULANASE SECRETION PROTEIN PULS | | 著者 | Tosi, T, Nickerson, N.N, Mollica, L, RingkjobingJensen, M, Blackledge, M, Baron, B, England, P, Pugsley, A.P, Dessen, A. | | 登録日 | 2011-10-24 | | 公開日 | 2011-12-07 | | 最終更新日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Pilotin-Secretin Recognition in the Type II Secretion System of Klebsiella Oxytoca.
Mol.Microbiol, 82, 2011
|
|
7UUR
 
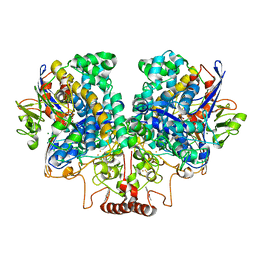 | | The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L) | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, HYDROXIDE ION, ... | | 著者 | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | 登録日 | 2022-04-28 | | 公開日 | 2023-01-04 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (1.67 Å) | | 主引用文献 | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UUS
 
 | | The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | 著者 | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | 登録日 | 2022-04-28 | | 公開日 | 2023-01-04 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
7UTD
 
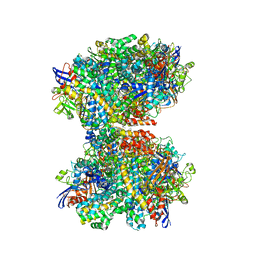 | | The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2, ... | | 著者 | Grinter, R, Venugopal, H, Kropp, A, Greening, C. | | 登録日 | 2022-04-26 | | 公開日 | 2023-01-04 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (2.19 Å) | | 主引用文献 | Structural basis for bacterial energy extraction from atmospheric hydrogen.
Nature, 615, 2023
|
|
1BHW
 
 | | LOW TEMPERATURE MIDDLE RESOLUTION STRUCTURE OF XYLOSE ISOMERASE FROM MASC DATA | | 分子名称: | XYLOSE ISOMERASE | | 著者 | Ramin, M, Shepard, W, Fourme, R, Kahn, R. | | 登録日 | 1998-06-10 | | 公開日 | 1998-11-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Multiwavelength anomalous solvent contrast (MASC): derivation of envelope structure-factor amplitudes and comparison with model values.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
