5RFY
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102974 | | 分子名称: | 1-acetyl-N-methyl-N-(propan-2-yl)piperidine-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4B1L
 
 | | CARBOHYDRATE BINDING MODULE CBM66 FROM BACILLUS SUBTILIS | | 分子名称: | LEVANASE, SODIUM ION, beta-D-fructofuranose | | 著者 | Cuskin, F, Flint, J.E, Morland, C, Basle, A, Henrissat, B, Countinho, P.M, Strazzulli, A, Solzehinkin, A, Davies, G.J, Gilbert, H.J, Gloster, T.M. | | 登録日 | 2012-07-11 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | How Nature Can Exploit Nonspecific Catalytic and Carbohydrate Binding Modules to Create Enzymatic Specificity
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5REI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2856434856 | | 分子名称: | 3C-like proteinase, 4-[(3-chlorophenyl)methyl]morpholine, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5REY
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102911 | | 分子名称: | 1-{4-[(2-methylphenyl)methyl]-1,4-diazepan-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFD
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z126932614 | | 分子名称: | 2-[(methylsulfonyl)methyl]-1H-benzimidazole, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFS
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102739 | | 分子名称: | 1-{4-[(thiophen-3-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RGR
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z328695024 (Mpro-x1101) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N,1-dimethyl-N-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-04-07 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3PAT
 
 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN | | 分子名称: | CALCIUM ION, PARVALBUMIN | | 著者 | Padilla, A, Cave, A, Parello, J, Etienne, G, Baldellon, C. | | 登録日 | 1994-03-22 | | 公開日 | 1994-07-31 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin
To be Published
|
|
4MZM
 
 | | MazF from S. aureus crystal form I, P212121, 2.1 A | | 分子名称: | mRNA interferase MazF | | 著者 | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | 登録日 | 2013-09-30 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
4J9C
 
 | |
4J9G
 
 | |
1XYE
 
 | | T-to-THigh Transitions in Human Hemoglobin: alpha Y42A deoxy low salt | | 分子名称: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Kavanaugh, J.S, Rogers, P.H, Arnone, A, Hui, H.L, Wierzba, A, DeYoung, A, Kwiatkowski, L.D, Noble, R.W, Juszczak, L.J, Peterson, E.S, Friedman, J.M. | | 登録日 | 2004-11-09 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Intersubunit interactions associated with tyr42alpha stabilize the quaternary-T tetramer but are not major quaternary constraints in deoxyhemoglobin
Biochemistry, 44, 2005
|
|
4JJD
 
 | |
1US0
 
 | | Human Aldose Reductase in complex with NADP+ and the inhibitor IDD594 at 0.66 Angstrom | | 分子名称: | ALDOSE REDUCTASE, CITRIC ACID, IDD594, ... | | 著者 | Howard, E.I, Sanishvili, R, Cachau, R.E, Mitschler, A, Chevrier, B, Barth, P, Lamour, V, Van Zandt, M, Sibley, E, Bon, C, Moras, D, Schneider, T.R, Joachimiak, A, Podjarny, A. | | 登録日 | 2003-11-16 | | 公開日 | 2004-05-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (0.66 Å) | | 主引用文献 | Ultrahigh Resolution Drug Design I: Details of Interactions in Human Aldose Reductase-Inhibitor Complex at 0.66 A.
Proteins, 55, 2004
|
|
1VLK
 
 | |
2L2D
 
 | | Solution NMR Structure of human UBA-like domain of OTUD7A_11_83, NESG target HT6304A/OCSP target OTUD7A_11_83/SGC-Toronto | | 分子名称: | OTU domain-containing protein 7A | | 著者 | Wu, B, Yee, A, Lemak, A, Gutmanas, A, Houliston, S, Semesi, A, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Structural Genomics Consortium (SGC) | | 登録日 | 2010-08-17 | | 公開日 | 2010-09-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The amino-terminal UBA domain of OTUD7A
To be Published
|
|
3PF3
 
 | | Crystal structure of a mutant (C202A) of Triosephosphate isomerase from Giardia lamblia derivatized with MMTS | | 分子名称: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | 著者 | Enriquez-Flores, S, Rodriguez-Romero, A, Hernandez-Santoyo, A, Reyes-Vivas, H. | | 登録日 | 2010-10-27 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | Determining the molecular mechanism of inactivation by chemical modification of triosephosphate isomerase from the human parasite Giardia lamblia: A study for antiparasitic drug design.
Proteins, 79, 2011
|
|
3TJY
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited chymotrypsin digestion | | 分子名称: | CHLORIDE ION, Effector protein hopAB3, SULFATE ION | | 著者 | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-08-25 | | 公開日 | 2011-09-14 | | 最終更新日 | 2013-01-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors.
Biochemistry, 51, 2012
|
|
2FVJ
 
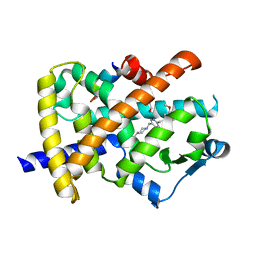 | | A novel anti-adipogenic partial agonist of peroxisome proliferator-activated receptor-gamma (PPARG) recruits pparg-coactivator-1 alpha (PGC1A) but potentiates insulin signaling in vitro | | 分子名称: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXY-4-{[4-(2-METHOXYPHENYL)PIPERIDIN-1-YL]METHYL}ISOQUINOLINE, GLYCEROL, Nuclear receptor coactivator 1, ... | | 著者 | Benz, J, Burgermeister, E, Flament, A, Schnoebelen, A, Stihle, M, Gsell, B, Rufer, A, Ruf, A, Kuhn, B, Maerki, H.P, Mizrahi, J, Sebokova, E, Niesor, E, Meyer, M. | | 登録日 | 2006-01-31 | | 公開日 | 2006-05-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | A novel partial agonist of peroxisome proliferator-activated receptor-gamma (PPARgamma) recruits PPARgamma-coactivator-1alpha, prevents triglyceride accumulation, and potentiates insulin signaling in vitro
Mol.Endocrinol., 20, 2006
|
|
6THZ
 
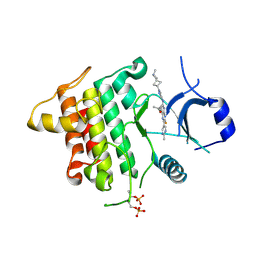 | | IRAK4 IN COMPLEX WITH inhibitor | | 分子名称: | 7-fluoranyl-~{N}-[1-(2-methyl-2-azaspiro[3.3]heptan-6-yl)pyrazol-4-yl]-4-(1-methylcyclopropyl)oxy-6-(2-methylpyrimidin-5-yl)pyrido[3,2-d]pyrimidin-2-amine, Interleukin-1 receptor-associated kinase 4 | | 著者 | Xue, Y, Aagaard, A, Degorce, S.L. | | 登録日 | 2019-11-21 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Improving metabolic stability and removing aldehyde oxidase liability in a 5-azaquinazoline series of IRAK4 inhibitors.
Bioorg.Med.Chem., 28, 2020
|
|
6TQ5
 
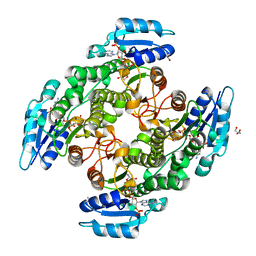 | | Alcohol dehydrogenase from Candida magnoliae DSMZ 70638 (ADHA): complex with NADP+ | | 分子名称: | Enzyme subunit, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Rovida, S, Aalbers, F.S, Fraaije, M.W, Mattevi, A. | | 登録日 | 2019-12-16 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Approaching boiling point stability of an alcohol dehydrogenase through computationally-guided enzyme engineering.
Elife, 9, 2020
|
|
1HRI
 
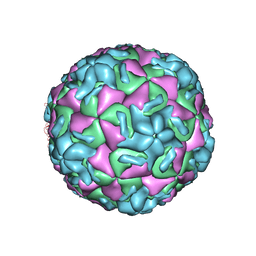 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | 分子名称: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | 著者 | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | 登録日 | 1992-10-01 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
6HO3
 
 | |
6HOC
 
 | |
5R82
 
 | | PanDDA analysis group deposition -- Crystal Structure of COVID-19 main protease in complex with Z219104216 | | 分子名称: | 3C-like proteinase, 6-(ethylamino)pyridine-3-carbonitrile, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Powell, A.J, Douangamath, A, Owen, C.D, Wild, C, Krojer, T, Lukacik, P, Strain-Damerell, C.M, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-03 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
