5MJ3
 
 | | INTERLEUKIN-23 COMPLEX WITH AN ANTAGONISTIC ALPHABODY, CRYSTAL FORM 1 | | 分子名称: | ALPHABODY MA12, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | 著者 | Desmet, J, Verstraete, K, Bloch, Y, Lorent, E, Wen, Y, Devreese, B, Vandenbroucke, K, Loverix, S, Hettmann, T, Deroo, S, Somers, K, Hendrikx, P, Lasters, I, Savvides, S.N. | | 登録日 | 2016-11-29 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural Basis Of Il-23 Antagonism By An Alphabody Protein Scaffold.
Nat Commun, 5, 2014
|
|
3BM9
 
 | | Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 | | 分子名称: | 4-bromo-6-(6-hydroxy-1,2-benzisoxazol-3-yl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | 著者 | Gopalsamy, A, Shi, M, Vogan, E.M, Golas, J, Jacob, J, Johnson, J, Lee, F, Nilakantan, R, Peterson, R, Svenson, K, Tam, M.S, Wen, Y, Chopra, R, Ellingboe, J, Arndt, K, Boschelli, F. | | 登録日 | 2007-12-12 | | 公開日 | 2008-07-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90.
J.Med.Chem., 51, 2008
|
|
3BMY
 
 | | Discovery of Benzisoxazoles as Potent Inhibitors of Chaperone Hsp90 | | 分子名称: | 4-chloro-6-{5-[(2-morpholin-4-ylethyl)amino]-1,2-benzisoxazol-3-yl}benzene-1,3-diol, Heat shock protein HSP 90-alpha | | 著者 | Gopalsamy, A, Shi, M, Vogan, E.M, Golas, J, Jacob, J, Johnson, J, Lee, F, Nilakantan, R, Peterson, R, Svenson, K, Tam, M.S, Wen, Y, Chopra, R, Ellingboe, J, Arndt, K, Boschelli, F. | | 登録日 | 2007-12-13 | | 公開日 | 2008-07-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of benzisoxazoles as potent inhibitors of chaperone heat shock protein 90.
J.Med.Chem., 51, 2008
|
|
2KDY
 
 | | NMR structure of LP2086-B01 | | 分子名称: | Factor H binding protein variant B01_001 | | 著者 | Mascioni, A, Bentley, B.E, Camarda, R, Dilts, D.A, Fink, P, Gusarova, V, Hoiseth, S, Jacob, J, Lin, S.L, Malakian, K, McNeil, L.K, Mininni, T, Moy, F, Murphy, E, Novikova, E, Sigethy, S, Wen, Y, Zlotnick, G.W, Tsao, D.H.H. | | 登録日 | 2009-01-20 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Immunogenic Properties of the Meningococcal Vaccine Candidate LP2086.
J.Biol.Chem., 284, 2009
|
|
6NSJ
 
 | | CryoEM structure of Helicobacter pylori urea channel in closed state | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | 著者 | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | 登録日 | 2019-01-24 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6NSK
 
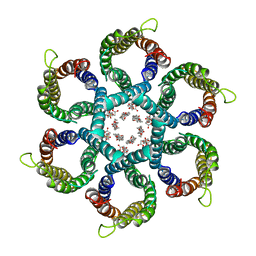 | | CryoEM structure of Helicobacter pylori urea channel in open state. | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | 著者 | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | 登録日 | 2019-01-24 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
2MUX
 
 | |
6KCZ
 
 | |
8IMW
 
 | |
7VPG
 
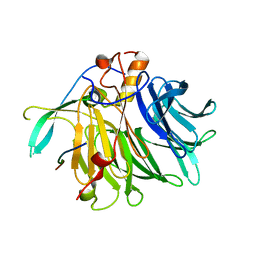 | | Crystal structure of the C-terminal tail of SARS-CoV-1 Orf6 complex with human nucleoporin pair Rae1-Nup98 | | 分子名称: | Isoform 3 of Nuclear pore complex protein Nup98-Nup96, ORF6 protein, mRNA export factor | | 著者 | Li, T, Guo, H, Yang, T, Wen, Y, Ji, X. | | 登録日 | 2021-10-17 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1-Nup98 Complex to Compete With mRNA Nuclear Export.
Front Mol Biosci, 8, 2021
|
|
7VPH
 
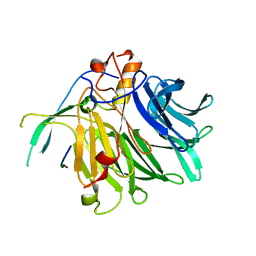 | | Crystal structure of the C-terminal tail of SARS-CoV-2 Orf6 complex with human nucleoporin pair Rae1-Nup98 | | 分子名称: | Isoform 3 of Nuclear pore complex protein Nup98-Nup96, ORF6 protein, mRNA export factor | | 著者 | Li, T, Guo, H, Yang, T, Wen, Y, Ji, X. | | 登録日 | 2021-10-17 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1-Nup98 Complex to Compete With mRNA Nuclear Export.
Front Mol Biosci, 8, 2021
|
|
3G60
 
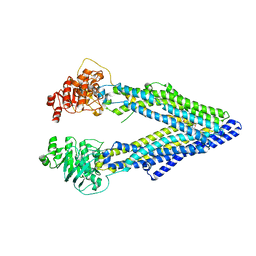 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | 分子名称: | (4R,11R,18R)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1a | | 著者 | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | 登録日 | 2009-02-05 | | 公開日 | 2009-03-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (4.4 Å) | | 主引用文献 | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
3G5U
 
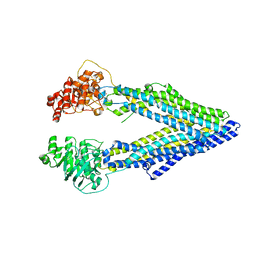 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | 分子名称: | MERCURY (II) ION, Multidrug resistance protein 1a | | 著者 | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | 登録日 | 2009-02-05 | | 公開日 | 2009-03-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
3G61
 
 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | 分子名称: | (4S,11S,18S)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1a | | 著者 | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | 登録日 | 2009-02-05 | | 公開日 | 2009-03-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (4.35 Å) | | 主引用文献 | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
3P50
 
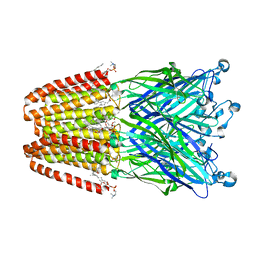 | | Structure of propofol bound to a pentameric ligand-gated ion channel, GLIC | | 分子名称: | 2,6-BIS(1-METHYLETHYL)PHENOL, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | 著者 | Nury, H, Van Renterghem, C, Weng, Y, Tran, A, Baaden, M, Dufresne, V, Changeux, J.P, Sonner, J.M, Delarue, M, Corringer, P.J. | | 登録日 | 2010-10-07 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel
Nature, 469, 2011
|
|
3P4W
 
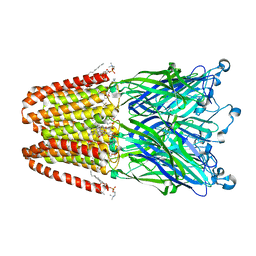 | | Structure of desflurane bound to a pentameric ligand-gated ion channel, GLIC | | 分子名称: | (2S)-2-(difluoromethoxy)-1,1,1,2-tetrafluoroethane, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | 著者 | Nury, H, Van Renterghem, C, Weng, Y, Tran, A, Baaden, M, Dufresne, V, Changeux, J.P, Sonner, J.M, Delarue, M, Corringer, P.J. | | 登録日 | 2010-10-07 | | 公開日 | 2011-01-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel
Nature, 469, 2011
|
|
2JOH
 
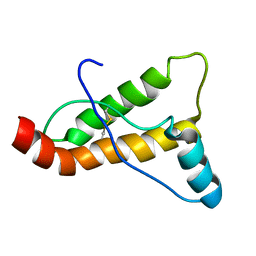 | |
2JOM
 
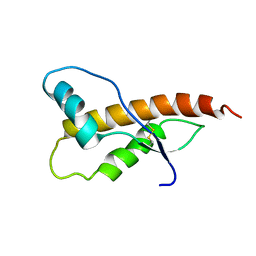 | |
2FJ3
 
 | |
5KZA
 
 | | Crystal structure of the Rous sarcoma virus matrix protein (aa 2-102). Space group I41 | | 分子名称: | 1,2-ETHANEDIOL, NITRATE ION, virus matrix protein | | 著者 | Kingston, R.L, Chan, J, Vogt, V.M. | | 登録日 | 2016-07-24 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Cholesterol Promotes Protein Binding by Affecting Membrane Electrostatics and Solvation Properties.
Biophys. J., 113, 2017
|
|
5KZ9
 
 | |
5KZB
 
 | |
6VJT
 
 | |
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | 著者 | Wang, K, Wang, X, Pan, L. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
