6F4Z
 
 | |
6FFR
 
 | |
6ERL
 
 | |
5OV2
 
 | |
5IWL
 
 | | CD47-diabody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5F9 diabody, Leukocyte surface antigen CD47, ... | | 著者 | Di, W, Jude, K.M, Garcia, K.C. | | 登録日 | 2016-03-22 | | 公開日 | 2016-06-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer.
J.Clin.Invest., 126, 2016
|
|
3S96
 
 | | Crystal structure of 3B5H10 | | 分子名称: | 3B5H10 FAB heavy chain, 3B5H10 FAB light chain | | 著者 | Weisgraber, K, Peters-Libeu, C, Rutenber, E, Newhouse, Y, Finkbeiner, S. | | 登録日 | 2011-05-31 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
4KJY
 
 | | Complex of high-affinity SIRP alpha variant FD6 with CD47 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, High-affinity SIRPa variant FD6, Leukocyte surface antigen CD47, ... | | 著者 | Ring, A.M, Ozkan, E, Ho, C.C.M, Garcia, K.C. | | 登録日 | 2013-05-04 | | 公開日 | 2013-06-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Engineered SIRP alpha variants as immunotherapeutic adjuvants to anticancer antibodies.
Science, 341, 2013
|
|
3V50
 
 | | Complex of SHV S130G mutant beta-lactamase complexed to SA2-13 | | 分子名称: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, Beta-lactamase, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | 著者 | Wei, K, van den Akker, F. | | 登録日 | 2011-12-15 | | 公開日 | 2012-08-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The importance of the trans-enamine intermediate as a beta-lactamase inhibition strategy probed in inhibitor-resistant SHV beta-lactamase variants.
Chemmedchem, 7, 2012
|
|
4KF7
 
 | |
4KF8
 
 | |
7SI9
 
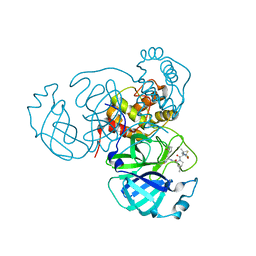 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with PF-07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-10-12 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
4DCQ
 
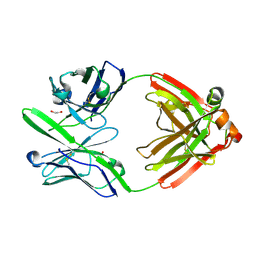 | | Crystal Structure of the Fab Fragment of 3B5H10, an Antibody-Specific for Extended Polyglutamine Repeats (orthorhombic form) | | 分子名称: | 1,2-ETHANEDIOL, 3B5H10 FAB Heavy Chain, 3B5H10 FAB Light Chain | | 著者 | Peters-Libeu, C.A, Tran, T, Finkbeiner, S, Weisgraber, K. | | 登録日 | 2012-01-18 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
6K9O
 
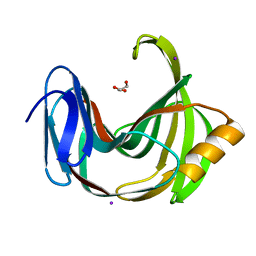 | | Crystal Structure Analysis of Protein | | 分子名称: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | 著者 | Li, C, Wan, Q. | | 登録日 | 2019-06-17 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
4RSG
 
 | | Neutron crystal structure of Ras bound to the GTP analogue GppNHp | | 分子名称: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Knihtila, R.R, Holzapfel, G, Weiss, K.L, Meilleur, F, Mattos, C. | | 登録日 | 2014-11-07 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | NEUTRON DIFFRACTION (1.907 Å) | | 主引用文献 | Neutron Crystal Structure of RAS GTPase Puts in Question the Protonation State of the GTP gamma-Phosphate.
J.Biol.Chem., 290, 2015
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | 分子名称: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-05 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | 分子名称: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-07 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-03 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-19 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-20 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-18 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-19 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5KSC
 
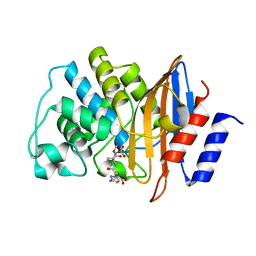 | | E166A/R274N/R276N Toho-1 Beta-lactamase aztreonam acyl-enzyme intermediate | | 分子名称: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Beta-lactamase Toho-1 | | 著者 | Vandavasi, V.G, Langan, P.S, Weiss, K, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | 登録日 | 2016-07-08 | | 公開日 | 2016-11-09 | | 最終更新日 | 2019-12-04 | | 実験手法 | NEUTRON DIFFRACTION (2.1 Å) | | 主引用文献 | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
4GD6
 
 | | SHV-1 beta-lactamase in complex with penam sulfone SA1-204 | | 分子名称: | (3R)-N-(2-formylindolizin-3-yl)-4-[(phenylacetyl)oxy]-3-sulfino-D-valine, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | 著者 | van den Akker, F, Wei, K. | | 登録日 | 2012-07-31 | | 公開日 | 2013-07-31 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Structures of SHV-1 beta-lactamase with penem and penam sulfone inhibitors that form cyclic intermediates stabilized by carbonyl conjugation
Plos One, 7, 2012
|
|
4HKL
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | 登録日 | 2012-10-15 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKO
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II (E177Q) in the apo form | | 分子名称: | Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | 登録日 | 2012-10-15 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
