3V50
 
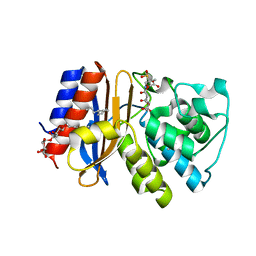 | | Complex of SHV S130G mutant beta-lactamase complexed to SA2-13 | | Descriptor: | (3R)-4-[(4-CARBOXYBUTANOYL)OXY]-N-[(1E)-3-OXOPROP-1-EN-1-YL]-3-SULFINO-D-VALINE, Beta-lactamase, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Wei, K, van den Akker, F. | | Deposit date: | 2011-12-15 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The importance of the trans-enamine intermediate as a beta-lactamase inhibition strategy probed in inhibitor-resistant SHV beta-lactamase variants.
Chemmedchem, 7, 2012
|
|
7LVM
 
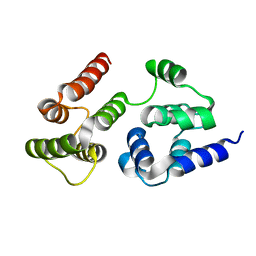 | |
7LVJ
 
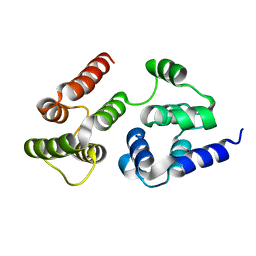 | | CASP8 isoform G DED domain | | Descriptor: | Isoform 9 of Caspase-8 | | Authors: | Weichert, K, Lu, F, Kodandapani, L, Sauder, J.M. | | Deposit date: | 2021-02-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caspase-8 Variant G Regulates Rheumatoid Arthritis Fibroblast-Like Synoviocyte Aggressive Behavior.
ACR Open Rheumatol, 4, 2022
|
|
1B6F
 
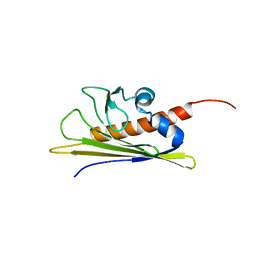 | | BIRCH POLLEN ALLERGEN BET V 1 | | Descriptor: | PROTEIN (MAJOR POLLEN ALLERGEN BET V 1-A) | | Authors: | Schweimer, K, Sticht, H, Boehm, M, Roesch, P. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Spectroscopy Reveals Common Structural Features of the Birch Pollen Allergen Bet v 1 and the cherry allergen Pru a 1
APPL.MAGN.RESON., 17, 1999
|
|
6YEP
 
 | |
6YCV
 
 | |
6R3C
 
 | |
1E0Z
 
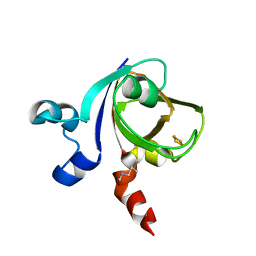 | | [2Fe-2S]-Ferredoxin from Halobacterium salinarum | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Schweimer, K, Marg, B, Oesterhelt, D, Roesch, P, Sticht, H. | | Deposit date: | 2000-04-11 | | Release date: | 2001-04-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | A Two-Alpha-Helix Extra Domain Mediates the Halophilic Character of a Plant-Type Ferredoxin from Halophilic Archaea.
Biochemistry, 44, 2005
|
|
1DX8
 
 | | Rubredoxin from Guillardia theta | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Schweimer, K, Hoffmann, S, Wastl, J, Maier, U.G, Roesch, P, Sticht, H. | | Deposit date: | 1999-12-23 | | Release date: | 2000-01-04 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a zinc substituted eukaryotic rubredoxin from the cryptomonad alga Guillardia theta.
Protein Sci., 9, 2000
|
|
1H7V
 
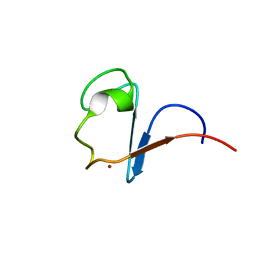 | | Rubredoxin from Guillardia Theta | | Descriptor: | RUBREDOXIN, ZINC ION | | Authors: | Schweimer, K, Hoffmann, S, Wastl, J, Maier, U.G, Roesch, P, Sticht, H. | | Deposit date: | 2001-01-16 | | Release date: | 2002-01-29 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a zinc substituted eukaryotic rubredoxin from the cryptomonad alga Guillardia theta.
Protein Sci., 9, 2000
|
|
1H92
 
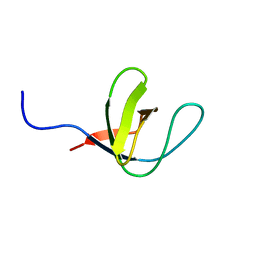 | | SH3 domain of human Lck tyrosine kinase | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Schweimer, K, Hoffmann, S, Friedrich, U, Biesinger, B, Roesch, P, Sticht, H. | | Deposit date: | 2001-02-22 | | Release date: | 2001-10-22 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural Investigation of the Binding of a Herpesviral Protein to the SH3 Domain of Tyrosine Kinase Lck
Biochemistry, 41, 2002
|
|
3S96
 
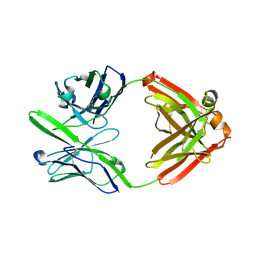 | | Crystal structure of 3B5H10 | | Descriptor: | 3B5H10 FAB heavy chain, 3B5H10 FAB light chain | | Authors: | Weisgraber, K, Peters-Libeu, C, Rutenber, E, Newhouse, Y, Finkbeiner, S. | | Deposit date: | 2011-05-31 | | Release date: | 2012-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
2BZ2
 
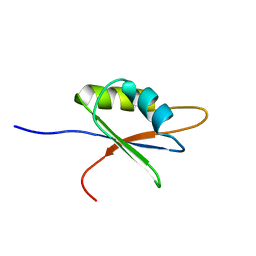 | | Solution structure of NELF E RRM | | Descriptor: | NEGATIVE ELONGATION FACTOR E | | Authors: | Schweimer, K, Rao, J.N, Neumann, L, Rosch, P, Wohrl, B.M. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-16 | | Last modified: | 2018-02-28 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the RNA-recognition motif of NELF E, a cellular negative transcription elongation factor involved in the regulation of HIV transcription.
Biochem. J., 400, 2006
|
|
2JVV
 
 | |
2K06
 
 | |
2JNZ
 
 | |
2KWP
 
 | |
4GD6
 
 | | SHV-1 beta-lactamase in complex with penam sulfone SA1-204 | | Descriptor: | (3R)-N-(2-formylindolizin-3-yl)-4-[(phenylacetyl)oxy]-3-sulfino-D-valine, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | van den Akker, F, Wei, K. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structures of SHV-1 beta-lactamase with penem and penam sulfone inhibitors that form cyclic intermediates stabilized by carbonyl conjugation
Plos One, 7, 2012
|
|
1XIP
 
 | |
7PER
 
 | | Model of the inner ring of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup155, Nuclear pore complex protein Nup205, Nuclear pore complex protein Nup93, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
1QGR
 
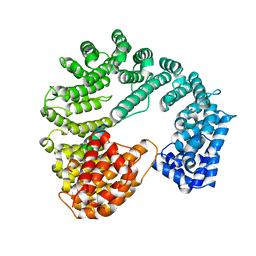 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA (II CRYSTAL FORM, GROWN AT LOW PH) | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-05-04 | | Release date: | 1999-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
1QGK
 
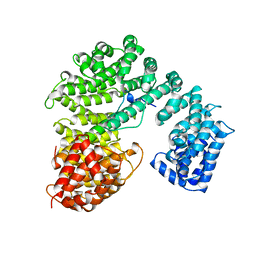 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
3GFP
 
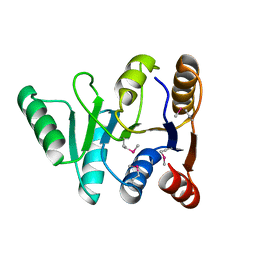 | | Structure of the C-terminal domain of the DEAD-box protein Dbp5 | | Descriptor: | DEAD box protein 5 | | Authors: | Erzberger, J.P, Dossani, Z.Y, Weirich, C.S, Weis, K, Berger, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3PEU
 
 | | S. cerevisiae Dbp5 L327V C-terminal domain bound to Gle1 H337R and IP6 | | Descriptor: | ATP-dependent RNA helicase DBP5, GLYCEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
