8EK5
 
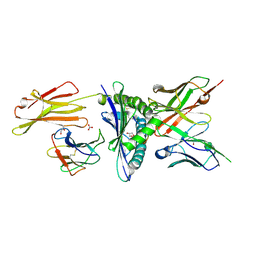 | | Engineered scFv 10LH bound to PHOX2B/HLA-A24:02 | | 分子名称: | 10LH single chain fragment variable (scFv), Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Garfinkle, S.E, Florio, T.J, Sgourakis, N.G. | | 登録日 | 2022-09-20 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
5VGU
 
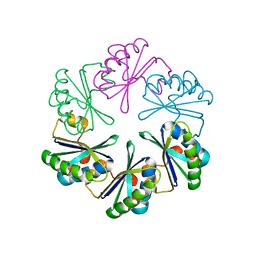 | |
4GMX
 
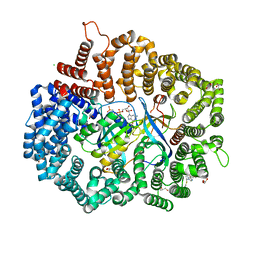 | |
8SBK
 
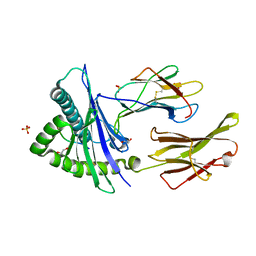 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI (ATG2A). | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, ... | | 著者 | Mallik, L, Young, M.C, Sgourakis, N.G. | | 登録日 | 2023-04-03 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
8SBL
 
 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI | | 分子名称: | Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, MHC class I antigen | | 著者 | Mallik, L, Young, M.C, Sgourakis, N.G. | | 登録日 | 2023-04-03 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
7BEM
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 scFv | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-269 Vh domain, ... | | 著者 | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2020-12-24 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEH
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-316 heavy chain, ... | | 著者 | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2020-12-23 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEN
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253 and COVOX-75 Fabs | | 分子名称: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | 著者 | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2020-12-24 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEO
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253H55L and COVOX-75 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2020-12-24 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEK
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 2) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2020-12-23 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEP
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-384 and S309 Fabs | | 分子名称: | CHLORIDE ION, COVOX-384 heavy chain, COVOX-384 light chain, ... | | 著者 | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2020-12-24 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEL
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-88 and COVOX-45 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COVOX-45 heavy chain, ... | | 著者 | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2020-12-23 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEI
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-150 heavy chain, ... | | 著者 | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2020-12-23 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEJ
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | 著者 | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2020-12-23 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
8EQI
 
 | |
4XB4
 
 | | Structure of the N-terminal domain of OCP binding canthaxanthin | | 分子名称: | Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | 著者 | Kerfeld, C.A, Sutter, M, Leverenz, R.L. | | 登録日 | 2014-12-16 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.544 Å) | | 主引用文献 | PHOTOSYNTHESIS. A 12 angstrom carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection.
Science, 348, 2015
|
|
4XB5
 
 | | Structure of orange carotenoid protein binding canthaxanthin | | 分子名称: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-carotene-4,4'-dione | | 著者 | Kerfeld, C.A, Sutter, M, Leverenz, R.L. | | 登録日 | 2014-12-16 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | PHOTOSYNTHESIS. A 12 angstrom carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection.
Science, 348, 2015
|
|
4ZBN
 
 | |
2K9U
 
 | |
4NI7
 
 | | Crystal structure of human interleukin 6 in complex with a modified nucleotide aptamer (SOMAMER SL1025) | | 分子名称: | Interleukin-6, SODIUM ION, SOMAmer SL1025 | | 著者 | Davies, D, Edwards, T, Gelinas, A, Jarvis, T, Clifton, M.C. | | 登録日 | 2013-11-05 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of interleukin-6 in complex with a modified nucleic Acid ligand.
J.Biol.Chem., 289, 2014
|
|
4NI9
 
 | | Crystal structure of human interleukin 6 in complex with a modified nucleotide aptamer (SOMAMER SL1025), FORM 2 | | 分子名称: | Interleukin-6, SODIUM ION, SOMAmer SL1025 | | 著者 | Davies, D, Edwards, T, Gelinas, A, Jarvis, T, Clifton, M.C. | | 登録日 | 2013-11-05 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of interleukin-6 in complex with a modified nucleic Acid ligand.
J.Biol.Chem., 289, 2014
|
|
4O7K
 
 | | Crystal structure of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor | | 分子名称: | CHLORIDE ION, ISOPROPYL ALCOHOL, PHOSPHATE ION, ... | | 著者 | Maindola, P, Goyal, P, Arulandu, A. | | 登録日 | 2013-12-25 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.748 Å) | | 主引用文献 | Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids
Nat Commun, 5, 2014
|
|
4OVB
 
 | | Crystal structure of Oncogenic Suppression Activity Protein - A Plasmid Fertility Inhibition Factor, Gold (I) Cyanide derivative | | 分子名称: | GLYCEROL, GOLD (I) CYANIDE ION, PHOSPHATE ION, ... | | 著者 | Maindola, P, Goyal, P, Arulandu, A. | | 登録日 | 2014-02-21 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.034 Å) | | 主引用文献 | Multiple enzymatic activities of ParB/Srx superfamily mediate sexual conflict among conjugative plasmids
Nat Commun, 5, 2014
|
|
1BMK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB218655 | | 分子名称: | 4-(FLUOROPHENYL)-1-CYCLOPROPYLMETHYL-5-(2-AMINO-4-PYRIMIDINYL)IMIDAZOLE, PROTEIN (MAP KINASE P38) | | 著者 | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | 登録日 | 1998-07-23 | | 公開日 | 1999-07-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BL7
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB220025 | | 分子名称: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | 著者 | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | 登録日 | 1998-07-23 | | 公開日 | 1999-07-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
