2ABZ
 
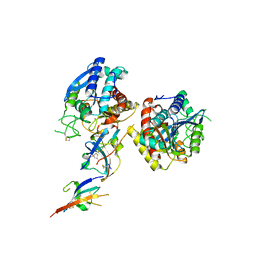 | | Crystal structure of C19A/C43A mutant of leech carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | 分子名称: | Carboxypeptidase A1, Metallocarboxypeptidase inhibitor, ZINC ION | | 著者 | Arolas, J.L, Popowicz, G.M, Bronsoms, S, Aviles, F.X, Huber, R, Holak, T.A, Ventura, S. | | 登録日 | 2005-07-18 | | 公開日 | 2006-01-31 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Study of a major intermediate in the oxidative folding of leech carboxypeptidase inhibitor: contribution of the fourth disulfide bond
J.Mol.Biol., 352, 2005
|
|
2DSQ
 
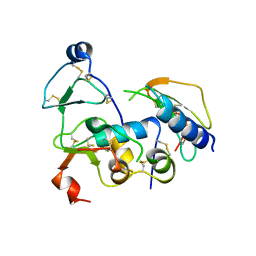 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | 分子名称: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 1, Insulin-like growth factor-binding protein 4 | | 著者 | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | 登録日 | 2006-07-05 | | 公開日 | 2006-08-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GRC
 
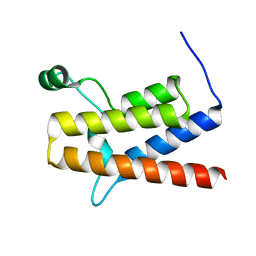 | | 1.5 A structure of bromodomain from human BRG1 protein, a central ATPase of SWI/SNF remodeling complex | | 分子名称: | Probable global transcription activator SNF2L4 | | 著者 | Singh, M, Popowicz, G.M, Krajewski, M, Holak, T.A. | | 登録日 | 2006-04-24 | | 公開日 | 2007-05-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural ramification for acetyl-lysine recognition by the bromodomain of human BRG1 protein, a central ATPase of the SWI/SNF remodeling complex.
Chembiochem, 8, 2007
|
|
2DSP
 
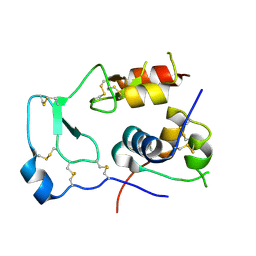 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | 分子名称: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 4 | | 著者 | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | 登録日 | 2006-07-05 | | 公開日 | 2006-08-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DSR
 
 | | Structural Basis for the Inhibition of Insulin-like Growth Factors by IGF Binding Proteins | | 分子名称: | Insulin-like growth factor IB, Insulin-like growth factor-binding protein 4 | | 著者 | Sitar, T, Popowicz, G.M, Siwanowicz, I, Huber, R, Holak, T.A. | | 登録日 | 2006-07-05 | | 公開日 | 2006-08-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1ZLI
 
 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with human carboxypeptidase B | | 分子名称: | Carboxypeptidase B, ZINC ION, carboxypeptidase inhibitor | | 著者 | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | 登録日 | 2005-05-06 | | 公開日 | 2005-07-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
1ZFI
 
 | | Solution structure of the leech carboxypeptidase inhibitor | | 分子名称: | Metallocarboxypeptidase inhibitor | | 著者 | Arolas, J.L, D'Silva, L, Popowicz, G.M, Aviles, F.X, Holak, T.A, Ventura, S. | | 登録日 | 2005-04-20 | | 公開日 | 2005-09-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structural characterization and computational predictions of the major intermediate in oxidative folding of leech carboxypeptidase inhibitor
STRUCTURE, 13, 2005
|
|
1ZFL
 
 | | Solution structure of III-A, the major intermediate in the oxidative folding of leech carboxypeptidase inhibitor | | 分子名称: | Metallocarboxypeptidase inhibitor | | 著者 | Arolas, J.L, D'Silva, L, Popowicz, G.M, Aviles, F.X, Holak, T.A, Ventura, S. | | 登録日 | 2005-04-20 | | 公開日 | 2005-09-13 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structural characterization and computational predictions of the major intermediate in oxidative folding of leech carboxypeptidase inhibitor
STRUCTURE, 13, 2005
|
|
1ZLH
 
 | | Crystal structure of the tick carboxypeptidase inhibitor in complex with bovine carboxypeptidase A | | 分子名称: | Carboxypeptidase A1, ZINC ION, carboxypeptidase inhibitor | | 著者 | Arolas, J.L, Popowicz, G.M, Lorenzo, J, Sommerhoff, C.P, Huber, R, Aviles, F.X, Holak, T.A. | | 登録日 | 2005-05-06 | | 公開日 | 2005-07-05 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Three-Dimensional Structures of Tick Carboxypeptidase Inhibitor in Complex with A/B Carboxypeptidases Reveal a Novel Double-headed Binding Mode
J.Mol.Biol., 350, 2005
|
|
1YZ5
 
 | |
4MDQ
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | 分子名称: | 3-[(1R)-2-(benzylamino)-1-{[(2S)-1-(hydroxyamino)-4-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl]-6-chloro-N-hydroxy-1H-indole-2-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Bista, M, Popowicz, G, Holak, T.A. | | 登録日 | 2013-08-23 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.119 Å) | | 主引用文献 | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
4MDN
 
 | | Structure of a novel submicromolar MDM2 inhibitor | | 分子名称: | 3-{(1S)-2-(tert-butylamino)-1-[{4-[(4-chlorobenzyl)oxy]benzyl}(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | 著者 | Bista, M, Popowicz, G, Holak, T.A. | | 登録日 | 2013-08-23 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.905 Å) | | 主引用文献 | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
8OS1
 
 | | X-ray structure of the Peroxisomal Targeting Signal 1 (PTS1) of Trypanosoma Cruzi PEX5 in complex with the PTS1 peptide | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Napolitano, V, Blat, A, Popowicz, G.M, Dubin, G. | | 登録日 | 2023-04-17 | | 公開日 | 2024-10-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural dynamics of the TPR domain of the peroxisomal cargo receptor Pex5 in Trypanosoma.
Int.J.Biol.Macromol., 280, 2024
|
|
3UFA
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | 分子名称: | CHLORIDE ION, N-(3-carboxypropanoyl)-L-valyl-N-[(1S)-2-phenyl-1-phosphonoethyl]-L-prolinamide, Serine protease splA | | 著者 | Zdzalik, M, Pietrusewicz, E, Pustelny, K, Stec-Niemczyk, J, Popowicz, G.M, Potempa, J, Oleksyszyn, J, Dubin, G. | | 登録日 | 2011-10-31 | | 公開日 | 2013-01-23 | | 最終更新日 | 2014-03-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
4MVN
 
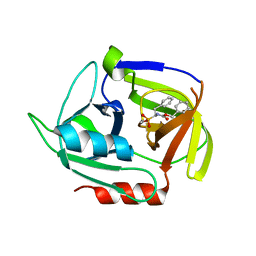 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | 分子名称: | Serine protease splA, [(1S)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl]phosphonic acid | | 著者 | Zdzalik, M, Burchacka, E, Niemczyk, J.S, Pustelny, K, Popowicz, G.M, Wladyka, B, Dubin, A, Potempa, J, Sienczyk, M, Dubin, G, Oleksyszyn, J. | | 登録日 | 2013-09-24 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
7R2V
 
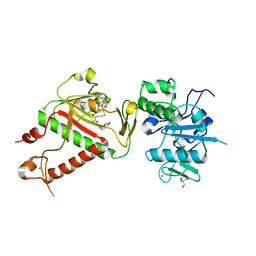 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | 著者 | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | 登録日 | 2022-02-06 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
8BW5
 
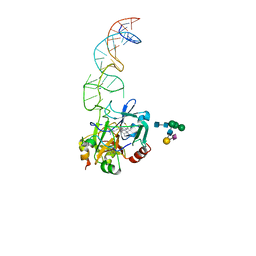 | | X-ray structure of the complex between human alpha thrombin and the duplex/quadruplex aptamer M08s-1_41mer | | 分子名称: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, M08s-1_41mer, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Troisi, R, Napolitano, V, Sica, F. | | 登録日 | 2022-12-06 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Steric hindrance and structural flexibility shape the functional properties of a guanine-rich oligonucleotide.
Nucleic Acids Res., 51, 2023
|
|
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | 分子名称: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | 登録日 | 2020-03-06 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
6R73
 
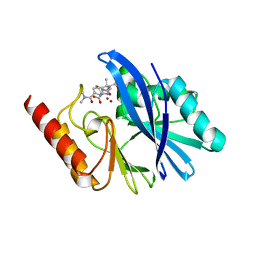 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed meropenem | | 分子名称: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, ZINC ION | | 著者 | Softley, C.A, Zak, K, Kolonko, M, Sattler, M, Popowicz, G. | | 登録日 | 2019-03-28 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6ZFW
 
 | | X-ray structure of the soluble N-terminal domain of T. cruzi PEX-14 | | 分子名称: | ACETATE ION, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | 著者 | Softley, C.A, Ostertag, M.O, Sattler, M, Popowicz, G.P. | | 登録日 | 2020-06-18 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Deep learning model predicts water interaction sites on the surface of proteins using limited-resolution data.
Chem.Commun.(Camb.), 56, 2020
|
|
5CH8
 
 | | Crystal structure of MDLA N225Q mutant form Penicillium cyclopium | | 分子名称: | CHLORIDE ION, GLYCEROL, Mono- and diacylglycerol lipase, ... | | 著者 | Xu, J, Xu, H, Hou, S, Liu, J. | | 登録日 | 2015-07-10 | | 公開日 | 2016-04-20 | | 最終更新日 | 2017-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Lipase-Driven Epoxidation Is A Two-Stage Synergistic Process
ChemistrySelect, 4, 2016
|
|
5MMC
 
 | |
7Q51
 
 | | yeast Gid10 bound to a Phe/N-peptide | | 分子名称: | CHLORIDE ION, FWLPANLW peptide, Uncharacterized protein YGR066C | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-11-02 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Multifaceted N-Degron Recognition and Ubiquitylation by GID/CTLH E3 Ligases.
J.Mol.Biol., 434, 2022
|
|
6GD2
 
 | | Structure of HuR RRM3 in complex with RNA | | 分子名称: | ELAV-like protein 1, RNA (5'-R(P*UP*UP*UP*AP*UP*UP*U)-3') | | 著者 | Pabis, M, Sattler, M. | | 登録日 | 2018-04-21 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs.
Nucleic Acids Res., 47, 2019
|
|
6GD3
 
 | |
