7X0D
 
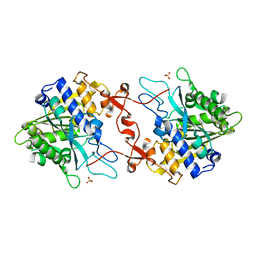 | | Crystal structure of phospholipase A1, CaPLA1 | | 分子名称: | Phospholipase A1, SULFATE ION | | 著者 | Heo, Y, Lee, I, Moon, S, Lee, W. | | 登録日 | 2022-02-21 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.39725161 Å) | | 主引用文献 | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
5G1E
 
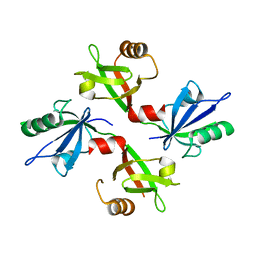 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | 分子名称: | SYNTENIN-1 | | 著者 | Lee, I, Kim, H, Yun, J.H, Lee, W. | | 登録日 | 2016-03-25 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
1MPV
 
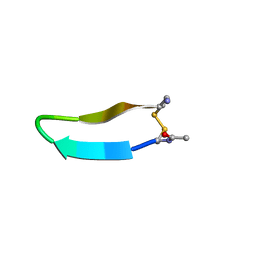 | | Structure of bhpBR3, the BAFF-binding loop of BR3 embedded in a beta-hairpin peptide | | 分子名称: | BLyS Receptor 3 | | 著者 | Kayagaki, N, Yan, M, Seshasayee, D, Wang, H, Lee, W, French, D.M, Grewal, I.S, Cochran, A.G, Gordon, N.C, Yin, J, Starovasnik, M.A, Dixit, V.M. | | 登録日 | 2002-09-12 | | 公開日 | 2002-10-30 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2.
Immunity, 17, 2002
|
|
1O07
 
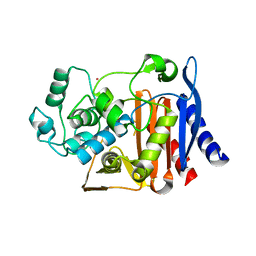 | | Crystal Structure of the complex between Q120L/Y150E mutant of AmpC and a beta-lactam inhibitor (MXG) | | 分子名称: | 2-(1-{2-[4-(2-ACETYLAMINO-PROPIONYLAMINO)-4-CARBOXY-BUTYRYLAMINO]-6-AMINO-HEXANOYLAMINO}-2-OXO-ETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, POTASSIUM ION | | 著者 | Meroueh, S.O, Minasov, G, Lee, W, Shoichet, B.K, Mobashery, S. | | 登録日 | 2003-02-20 | | 公開日 | 2003-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural Aspects for Evolution of beta-Lactamases from Penicillin-Binding Proteins
J.Am.Chem.Soc., 125, 2003
|
|
1L3X
 
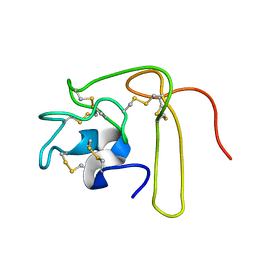 | |
1R02
 
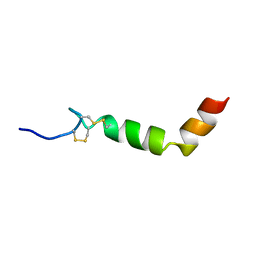 | |
1FUW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF A DOUBLE MUTANT SINGLE-CHAIN MONELLIN(SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | 分子名称: | MONELLIN | | 著者 | Sung, Y.H, Shin, J, Jung, J, Lee, W. | | 登録日 | 2000-09-18 | | 公開日 | 2001-06-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure, backbone dynamics, and stability of a double mutant single-chain monellin. structural origin of sweetness.
J.Biol.Chem., 276, 2001
|
|
1YY6
 
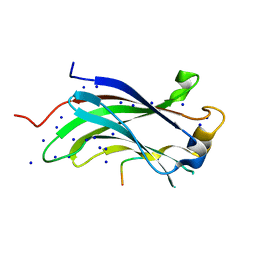 | | The Crystal Structure of the N-terminal domain of HAUSP/USP7 complexed with an EBNA1 peptide | | 分子名称: | Epstein-Barr nuclear antigen-1, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M, Shire, K, Nguyen, T, Zhang, R, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | 登録日 | 2005-02-23 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
1YZE
 
 | | Crystal structure of the N-terminal domain of USP7/HAUSP. | | 分子名称: | Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M.N, Shire, K, Nguyen, T, Zhang, R.G, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | 登録日 | 2005-02-28 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
1P5A
 
 | | Conformational Mapping of the N-terminal Peptide of HIV-1 GP41 in lipid detergent and aqueous environments using 13C-enhanced Fourier Transform Infrared Spectroscopy | | 分子名称: | Envelope polyprotein GP160 | | 著者 | Gordon, L.M, Mobley, P.W, Lee, W, Eskandari, S, Kaznessis, Y.N, Sherman, M.A, Waring, A.J. | | 登録日 | 2003-04-25 | | 公開日 | 2003-05-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | INFRARED SPECTROSCOPY | | 主引用文献 | Conformational mapping of the N-terminal peptide of HIV-1 gp41 in lipid detergent and aqueous environments using 13C-enhanced Fourier transform infrared spectroscopy.
Protein Sci., 13, 2004
|
|
1WPI
 
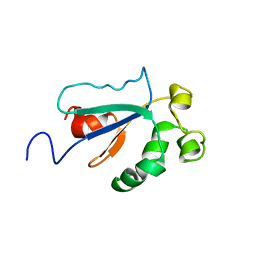 | | Solution NMR Structure of Protein YKR049C from Saccharomyces cerevisiae. Ontario Centre for Structural Proteomics target YST0250_1_133; Northeast Structural Genomics Consortium YTYst250 | | 分子名称: | Hypothetical 15.6 kDa protein in NAP1-TRK2 intergenic region | | 著者 | Jung, J.W, Yee, A, Arrowsmith, C.H, Lee, W, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-09-03 | | 公開日 | 2005-09-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of YKR049C, a putative redox protein from Saccharomyces cerevisiae
J.Biochem.Mol.Biol., 38, 2005
|
|
2ABY
 
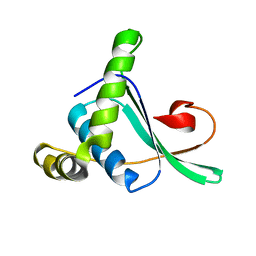 | | Solution structure of TA0743 from Thermoplasma acidophilum | | 分子名称: | hypothetical protein TA0743 | | 著者 | Kim, B, Jung, J, Hong, E, Yee, A, Arrowsmith, C.H, Lee, W. | | 登録日 | 2005-07-18 | | 公開日 | 2006-08-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the conserved novel-fold protein TA0743 from Thermoplasma acidophilum.
Proteins, 62, 2006
|
|
8WWX
 
 | |
6JCF
 
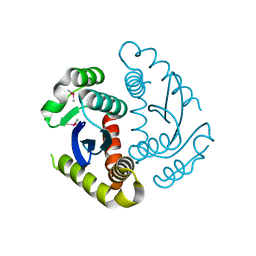 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | 分子名称: | CACODYLATE ION, Integrase | | 著者 | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | 登録日 | 2019-01-28 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.153 Å) | | 主引用文献 | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
1RQ6
 
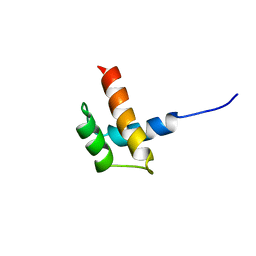 | | Solution structure of ribosomal protein S17E from Methanobacterium Thermoautotrophicum, Northeast Structural Genomics Consortium Target TT802 / Ontario Center for Structural Proteomics Target Mth0803 | | 分子名称: | 30S ribosomal protein S17e | | 著者 | Wu, B, Yee, A, Huang, Y.J, Ramelot, T.A, Semesi, A, Jung, J.W, Edward, A, Lee, W, Kennedy, M.A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-12-04 | | 公開日 | 2004-12-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of ribosomal protein S17E from Methanobacterium thermoautotrophicum: a structural homolog of the FF domain.
Protein Sci., 17, 2008
|
|
6JYA
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYF
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYE
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY7
 
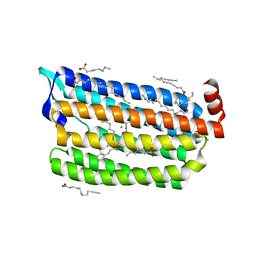 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYD
 
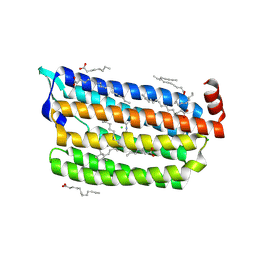 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.007 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY9
 
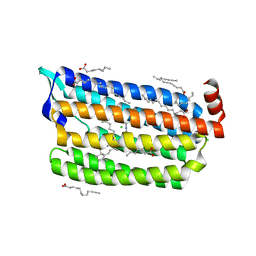 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYB
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
3K9O
 
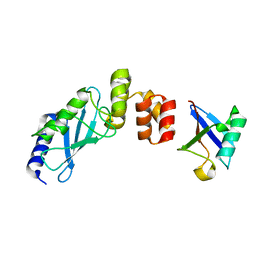 | | The crystal structure of E2-25K and UBB+1 complex | | 分子名称: | Ubiquitin, Ubiquitin-conjugating enzyme E2 K | | 著者 | Kang, G.B, Ko, S, Song, S.M, Lee, W, Eom, S.H. | | 登録日 | 2009-10-16 | | 公開日 | 2010-09-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of E2-25K/UBB+1 Interaction for Neurotoxicity of Alzheimer Disease by Proteasome Inhibition
To be Published
|
|
6JY6
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYC
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
