3T8A
 
 | | Crystal structure of Mycobacterium tuberculosis MenB in complex with substrate analogue, OSB-NCoA | | 分子名称: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, o-succinylbenzoyl-N-coenzyme A | | 著者 | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | 登録日 | 2011-08-01 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.245 Å) | | 主引用文献 | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
3T89
 
 | | Crystal structure of Escherichia coli MenB, the 1,4-dihydroxy-2-naphthoyl-CoA synthase in vitamin K2 biosynthesis | | 分子名称: | 1,4-Dihydroxy-2-naphthoyl-CoA synthase, GLYCEROL, MALONATE ION | | 著者 | Li, H.-J, Li, X, Liu, N, Zhang, H, Truglio, J, Mishra, S, Kisker, C, Garcia-Diaz, M, Tonge, P. | | 登録日 | 2011-08-01 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Mechanism of the Intramolecular Claisen Condensation Reaction Catalyzed by MenB, a Crotonase Superfamily Member.
Biochemistry, 50, 2011
|
|
6FDU
 
 | | Structure of Chlamydia trachomatis effector protein Cdu1 bound to Compound 3 | | 分子名称: | (2~{S},3~{S})-2-[[(2~{S})-2-[3,5-bis(chloranyl)phenyl]-2-(dimethylamino)ethanoyl]amino]-~{N}-[[2-(iminomethyl)pyrimidin-4-yl]methyl]-3-methyl-pentanamide, CHLORIDE ION, Deubiquitinase and deneddylase Dub1 | | 著者 | Ramirez, Y, Kisker, C, Altmann, E. | | 登録日 | 2017-12-26 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis of Substrate Recognition and Covalent Inhibition of Cdu1 from Chlamydia trachomatis.
ChemMedChem, 13, 2018
|
|
6FDQ
 
 | |
6FDK
 
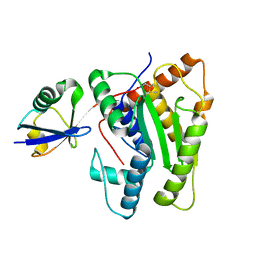 | |
3ZU5
 
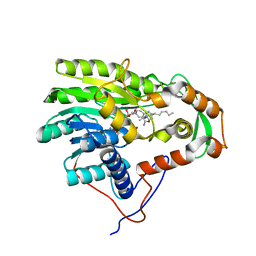 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT173 | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-hexylpyridin-2(1H)-one, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | 著者 | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
5LY9
 
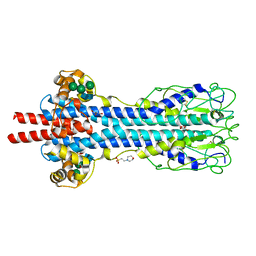 | | Structure of MITat 1.1 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Variant surface glycoprotein MITAT 1.1, ... | | 著者 | Schaefer, C, Bartossek, T, Jones, N, Kuper, J, Kisker, C, Engstler, M. | | 登録日 | 2016-09-26 | | 公開日 | 2017-09-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for the shielding function of the dynamic trypanosome variant surface glycoprotein coat.
Nat Microbiol, 2, 2017
|
|
3ZU3
 
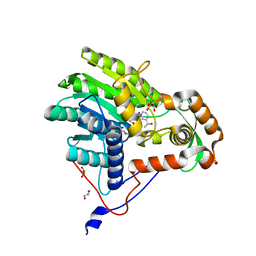 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (MR, cleaved Histag) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | 著者 | Hirschbeck, M.W, Kuper, J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU2
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (SIRAS) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, SODIUM ION | | 著者 | Hirschbeck, M.W, Kuper, J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
1RGC
 
 | | THE COMPLEX BETWEEN RIBONUCLEASE T1 AND 3'-GUANYLIC ACID SUGGESTS GEOMETRY OF ENZYMATIC REACTION PATH. AN X-RAY STUDY | | 分子名称: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | 著者 | Heydenreich, A, Koellner, G, Choe, H.W, Cordes, F, Kisker, C, Schindelin, H, Adamiak, R, Hahn, U, Saenger, W. | | 登録日 | 1993-05-12 | | 公開日 | 1994-01-31 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The complex between ribonuclease T1 and 3'GMP suggests geometry of enzymic reaction path. An X-ray study.
Eur.J.Biochem., 218, 1993
|
|
1T5L
 
 | | Crystal structure of the DNA repair protein UvrB point mutant Y96A revealing a novel fold for domain 2 | | 分子名称: | UvrABC system protein B, ZINC ION | | 著者 | Truglio, J.J, Croteau, D.L, Skorvaga, M, DellaVecchia, M.J, Theis, K, Mandavilli, B.S, Van Houten, B, Kisker, C. | | 登録日 | 2004-05-04 | | 公開日 | 2004-06-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Interactions between UvrA and UvrB: the role of UvrB's domain 2 in nucleotide excision repair
Embo J., 23, 2004
|
|
3ZU4
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT172 | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(2-CHLOROBENZYL)-4-HEXYLPYRIDIN-2(1H)-ONE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | 著者 | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | 登録日 | 2011-07-13 | | 公開日 | 2012-01-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
1Q52
 
 | | Crystal Structure of Mycobacterium tuberculosis MenB, a Key Enzyme in Vitamin K2 Biosynthesis | | 分子名称: | menB | | 著者 | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2003-08-05 | | 公開日 | 2004-01-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
5I4Z
 
 | | Structure of apo OmoMYC | | 分子名称: | CHLORIDE ION, GLYCEROL, Myc proto-oncogene protein, ... | | 著者 | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | 登録日 | 2016-02-13 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5I7S
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT01 | | 分子名称: | 5-ETHYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | 登録日 | 2016-02-18 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.595 Å) | | 主引用文献 | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I50
 
 | | Structure of OmoMYC bound to double-stranded DNA | | 分子名称: | DNA (5'-D(P*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*CP*TP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*CP*GP*GP*GP*TP*G)-3'), Myc proto-oncogene protein | | 著者 | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | 登録日 | 2016-02-13 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5I7F
 
 | |
5I8W
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT401 | | 分子名称: | 4-fluoro-5-hexyl-2-(2-methylphenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | 登録日 | 2016-02-19 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.629 Å) | | 主引用文献 | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I9N
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT412 | | 分子名称: | 5-ethyl-4-fluoro-2-(2-nitrophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | 登録日 | 2016-02-20 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.512 Å) | | 主引用文献 | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I8Z
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT12 | | 分子名称: | 5-HEXYL-2-(4-NITROPHENOXY)PHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | 登録日 | 2016-02-19 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.623 Å) | | 主引用文献 | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5IFL
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and triclosan | | 分子名称: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | 著者 | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | 登録日 | 2016-02-26 | | 公開日 | 2017-03-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
5I7E
 
 | |
5I9M
 
 | | Crystal structure of B. pseudomallei FabI in complex with NAD and PT408 | | 分子名称: | 5-ethyl-4-fluoro-2-[(2-methylpyridin-3-yl)oxy]phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Hirschbeck, M.W, Eltschkner, S, Tonge, P.J, Kisker, C. | | 登録日 | 2016-02-20 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Rationalizing the Binding Kinetics for the Inhibition of the Burkholderia pseudomallei FabI1 Enoyl-ACP Reductase.
Biochemistry, 56, 2017
|
|
1QRF
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | 分子名称: | CARBONIC ANHYDRASE, COBALT (II) ION, SULFATE ION | | 著者 | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | 登録日 | 1999-06-13 | | 公開日 | 1999-06-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRM
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | 分子名称: | CARBONIC ANHYDRASE, SULFATE ION, ZINC ION | | 著者 | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | 登録日 | 1999-06-15 | | 公開日 | 1999-06-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
