6LHC
 
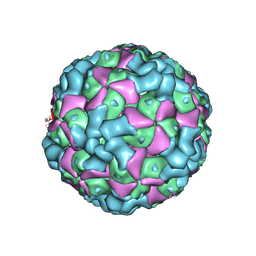 | | The cryo-EM structure of coxsackievirus A16 empty particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.43 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
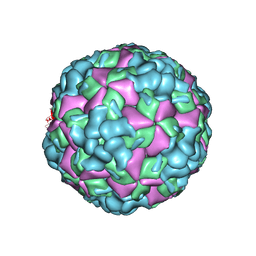 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | 分子名称: | VP1 protein, VP2 protein, VP3 protein | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHB
 
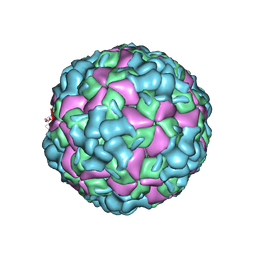 | | The cryo-EM structure of coxsackievirus A16 A-particle | | 分子名称: | VP1, VP2, VP3 | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHP
 
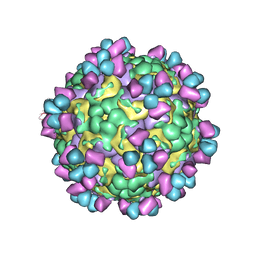 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 14B10 | | 分子名称: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | 著者 | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | 登録日 | 2019-12-09 | | 公開日 | 2020-02-05 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
7DPF
 
 | |
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | 分子名称: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | 分子名称: | VP2, VP3, Virion protein 1 | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | 登録日 | 2020-12-18 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | 分子名称: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | 著者 | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | 登録日 | 2020-12-22 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
4N6J
 
 | | Crystal structure of human Striatin-3 coiled coil domain | | 分子名称: | Striatin-3 | | 著者 | Chen, C, Shi, Z, Zhou, Z. | | 登録日 | 2013-10-13 | | 公開日 | 2014-02-26 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2:2 heterotetrameric core of striatin-interacting phosphatase and kinase (STRIPAK) complex.
J.Biol.Chem., 289, 2014
|
|
6LB0
 
 | |
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | 分子名称: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | 著者 | Xu, Z.H, Zhou, Z. | | 登録日 | 2020-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
8QNZ
 
 | | Crystal Structure of a Class D Carbapenemase Complexed with Hydrolyzed Imipenem | | 分子名称: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, 1-BUTANOL, BROMIDE ION, ... | | 著者 | Zhou, Q, He, Y, Jin, Y. | | 登録日 | 2023-09-27 | | 公開日 | 2023-11-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity.
Acs Cent.Sci., 9, 2023
|
|
4ZSE
 
 | |
6ZBO
 
 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in Complex with 1-(6-morpholinopyrimidin-4-yl)-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-5-ol (Molidustat) | | 分子名称: | 2-(6-morpholin-4-ylpyrimidin-4-yl)-4-(1,2,3-triazol-1-yl)pyrazol-3-ol, CHLORIDE ION, Egl nine homolog 1, ... | | 著者 | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Holt-Martyn, J.P, Schofield, C.J. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
6ZBN
 
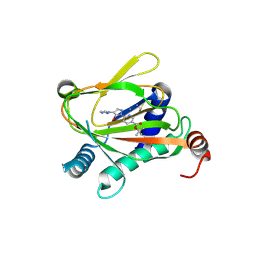 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in complex with tert-butyl 6-(5-hydroxy-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-1-yl)nicotinate (IOX4) | | 分子名称: | Egl nine homolog 1, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Schofield, C.J. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
8Y8E
 
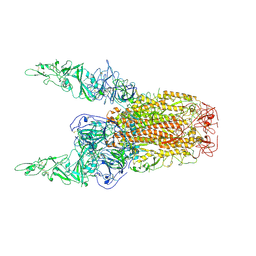 | | Structure of HCoV-HKU1C spike in the inactive-2up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8XSJ
 
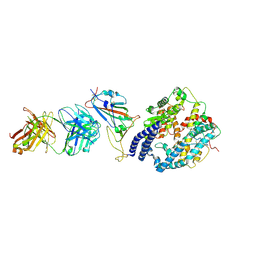 | | SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSE
 
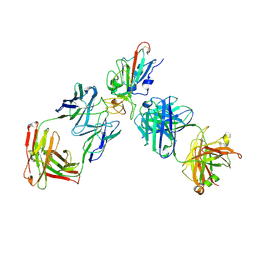 | | SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 H chain, IMCAS-123 L chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSF
 
 | | SARS-CoV-2 RBD + IMCAS-364 + hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, IMCAS-364 H chain, ... | | 著者 | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | 登録日 | 2024-01-09 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.16 Å) | | 主引用文献 | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8Y88
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-2up conformation with 2TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8Y7X
 
 | | Structure of HCoV-HKU1A spike in the functionally anchored-3up conformation with 3TMPRSS2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-05 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8Y8G
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-1up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
8Y8B
 
 | | Local structure of HCoV-HKU1C spike in complex with TMPRSS2 and glycan | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Wang, H.F, Zhang, X, Lu, Y.C, Liu, X.C, Sun, L, Yang, H.T. | | 登録日 | 2024-02-06 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.01 Å) | | 主引用文献 | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 2024
|
|
