5WXM
 
 | | Crystal structure of the Imp3 and Mpp10 complex | | 分子名称: | SULFATE ION, U3 small nucleolar RNA-associated protein MPP10, U3 small nucleolar ribonucleoprotein protein IMP3 | | 著者 | Ye, K, Zheng, S. | | 登録日 | 2017-01-07 | | 公開日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.304 Å) | | 主引用文献 | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5WYJ
 
 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | 著者 | Ye, K, Zhu, X, Sun, Q. | | 登録日 | 2017-01-13 | | 公開日 | 2017-03-29 | | 最終更新日 | 2019-10-09 | | 実験手法 | ELECTRON MICROSCOPY (8.7 Å) | | 主引用文献 | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WWM
 
 | |
5WYL
 
 | |
5I1W
 
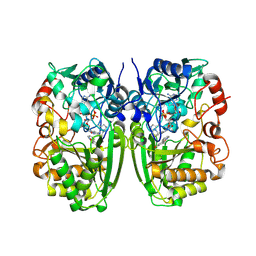 | | Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis | | 分子名称: | 4-hydroxy[2,2'-bipyridine]-6-carbaldehyde, 6-(hydroxymethyl)[2,2'-bipyridin]-4-ol, CrmK, ... | | 著者 | Picard, M.-E, Barma, J, Shi, R. | | 登録日 | 2016-02-07 | | 公開日 | 2017-02-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Biochemical and structural insights into flavoenzyme CrmK reveals a shunt product recycling mechanism in caerulomycin biosynthesis
to be published
|
|
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | 著者 | Wu, Y, Qi, J, Gao, F. | | 登録日 | 2020-04-26 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
7DF4
 
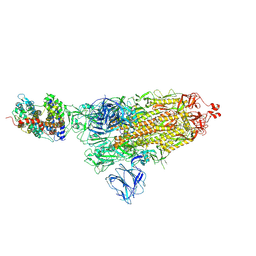 | | SARS-CoV-2 S-ACE2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Cong, X, Yao, C. | | 登録日 | 2020-11-06 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DF3
 
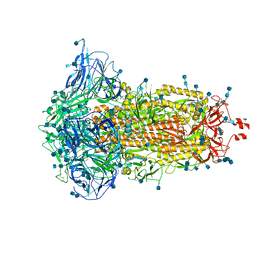 | | SARS-CoV-2 S trimer, S-closed | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cong, X, Yao, C. | | 登録日 | 2020-11-06 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7F1R
 
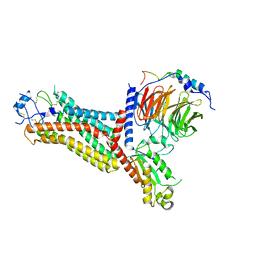 | | Cryo-EM structure of the chemokine receptor CCR5 in complex with RANTES and Gi | | 分子名称: | C-C motif chemokine 5,C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | 登録日 | 2021-06-09 | | 公開日 | 2021-07-14 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1T
 
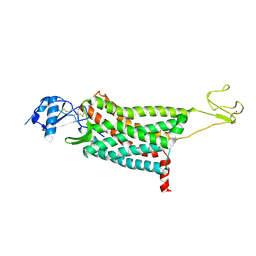 | | Crystal structure of the human chemokine receptor CCR5 in complex with MIP-1a | | 分子名称: | C-C motif chemokine 3,C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ZINC ION | | 著者 | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | 登録日 | 2021-06-09 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1S
 
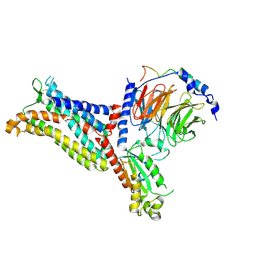 | | Cryo-EM structure of the apo chemokine receptor CCR5 in complex with Gi | | 分子名称: | C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | 登録日 | 2021-06-09 | | 公開日 | 2021-07-14 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1Q
 
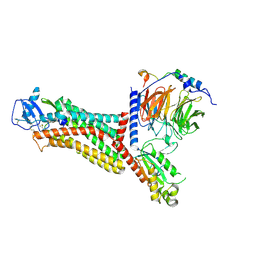 | | Cryo-EM structure of the chemokine receptor CCR5 in complex with MIP-1a and Gi | | 分子名称: | C-C motif chemokine 3,C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | 登録日 | 2021-06-09 | | 公開日 | 2021-07-14 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
6Z3Y
 
 | |
6A3N
 
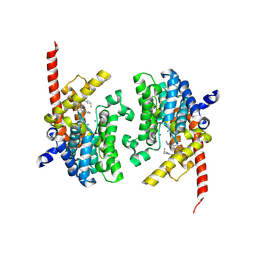 | | Crystal structure of the PDE9 catalytic domain in complex with inhibitor 2 | | 分子名称: | 1-cyclopentyl-6-({(2R)-1-[(3S)-3-fluoropyrrolidin-1-yl]-1-oxopropan-2-yl}amino)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Wu, Y.N, Zhou, Q, Chen, Y.P, Luo, H.B. | | 登録日 | 2018-06-15 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors against Phosphodiesterase-9, a Novel Target for the Treatment of Vascular Dementia.
J. Med. Chem., 62, 2019
|
|
1WMA
 
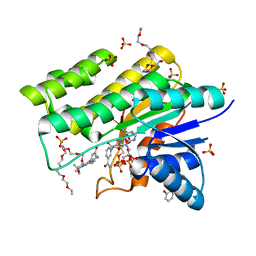 | | Crystal structure of human CBR1 in complex with Hydroxy-PP | | 分子名称: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, ... | | 著者 | Rauh, D, Bateman, R, Shokat, K.M. | | 登録日 | 2004-07-06 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | An unbiased cell morphology-based screen for new, biologically active small molecules
Plos Biol., 3, 2005
|
|
8K98
 
 | |
8K9A
 
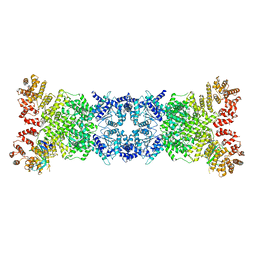 | | Cryo-EM structure of DSR2-DSAD1 state 2 | | 分子名称: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | 著者 | Zhang, H, Li, Z, Li, X.Z. | | 登録日 | 2023-07-31 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
7DNU
 
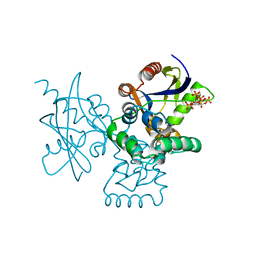 | | mRNA-decapping enzyme g5Rp with inhibitor insp6 complex | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, mRNA-decapping protein g5R | | 著者 | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | 登録日 | 2020-12-10 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.245 Å) | | 主引用文献 | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7DNT
 
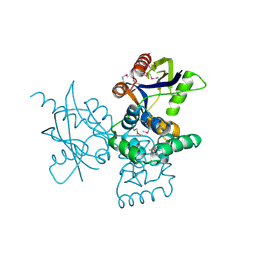 | | mRNA-decapping enzyme g5Rp | | 分子名称: | mRNA-decapping protein g5R | | 著者 | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | 登録日 | 2020-12-10 | | 公開日 | 2022-03-09 | | 最終更新日 | 2022-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
6ASG
 
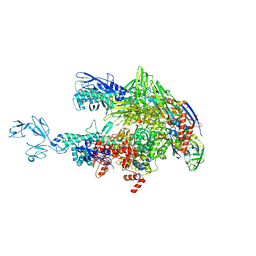 | | Crystal structure of Thermus thermophilus RNA polymerase core enzyme | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Liu, Y, Lin, W, Ying, R, Ebright, R.H. | | 登録日 | 2017-08-24 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
7DK3
 
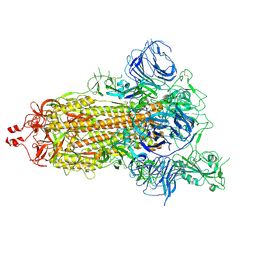 | | SARS-CoV-2 S trimer, S-open | | 分子名称: | Spike glycoprotein | | 著者 | Xu, C, Cong, Y. | | 登録日 | 2020-11-23 | | 公開日 | 2020-12-16 | | 最終更新日 | 2021-02-17 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
6Z3Z
 
 | |
7WUQ
 
 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | 分子名称: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | He, Q.T, Guo, S.C, Xiao, P, Sun, J.P, Yu, X, Gou, L, Kong, L.L, Zhang, L. | | 登録日 | 2022-02-09 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
3FM9
 
 | | Analysis of the Structural Determinants Underlying Discrimination between Substrate and Solvent in beta-Phosphoglucomutase Catalysis | | 分子名称: | Beta-phosphoglucomutase, MAGNESIUM ION | | 著者 | Finci, L, Lahiri, S, Peisach, E, Allen, K.N. | | 登録日 | 2008-12-19 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Analysis of the structural determinants underlying discrimination between substrate and solvent in beta-phosphoglucomutase catalysis.
Biochemistry, 48, 2009
|
|
7YSX
 
 | | Crystal structure of PDE4D complexed with licoisoflavone A | | 分子名称: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | 著者 | Liu, J.Y, Li, M.J, Xu, Y.C. | | 登録日 | 2022-08-13 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
