8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | 分子名称: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | 登録日 | 2022-09-08 | | 公開日 | 2022-11-09 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
6L4R
 
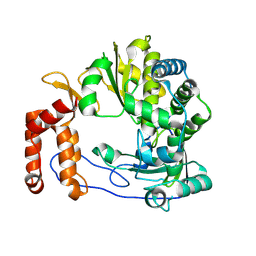 | | Crystal structure of Enterovirus D68 RdRp | | 分子名称: | RdRp | | 著者 | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | 登録日 | 2019-10-21 | | 公開日 | 2020-06-10 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (2.147 Å) | | 主引用文献 | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
8T5C
 
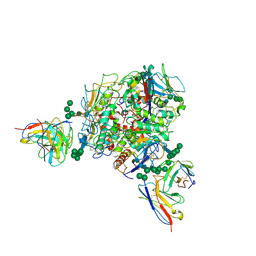 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | 著者 | Gorman, J, Kwong, P.D. | | 登録日 | 2023-06-13 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
7TVA
 
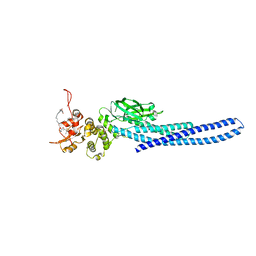 | | Stat5a Core in complex with AK2292 | | 分子名称: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2022-02-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.835 Å) | | 主引用文献 | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7TVB
 
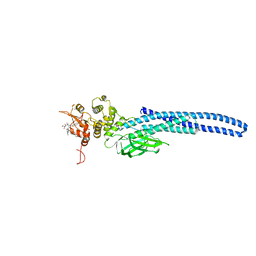 | | Stat5A Core in Complex with AK305 | | 分子名称: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2022-02-04 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.653 Å) | | 主引用文献 | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
1SN1
 
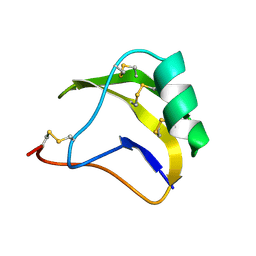 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | 分子名称: | PROTEIN (NEUROTOXIN BMK M1) | | 著者 | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | 登録日 | 1998-11-12 | | 公開日 | 1999-11-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1CHZ
 
 | | A NEW NEUROTOXIN FROM BUTHUS MARTENSII KARSCH | | 分子名称: | CHLORIDE ION, PROTEIN (BMK M2) | | 著者 | He, X.L, Deng, J.P, Li, H.M, Wang, D.C. | | 登録日 | 1999-03-31 | | 公開日 | 2000-03-31 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structure of a new neurotoxin from the scorpion Buthus martensii Karsch at 1.76 A.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1T0Z
 
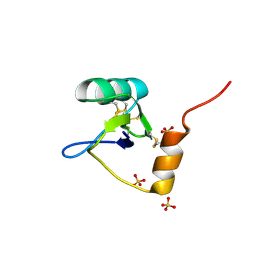 | | Structure of an Excitatory Insect-specific Toxin with an Analgesic Effect on Mammalian from Scorpion Buthus martensii Karsch | | 分子名称: | SULFATE ION, insect neurotoxin | | 著者 | Li, C, Guan, R.-J, Xiang, Y, Zhang, Y, Wang, D.-C. | | 登録日 | 2004-04-14 | | 公開日 | 2004-12-28 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of an excitatory insect-specific toxin with an analgesic effect on mammals from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2I1T
 
 | |
6WMW
 
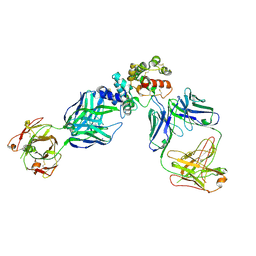 | | GFRAL receptor bound with two antibody Fabs (3P10, 25M22) | | 分子名称: | FAB25M22 heavy chain fragment, FAB25M22 light chain, FAB3P10 heavy chain fragment, ... | | 著者 | White, A, Lakshminarasimhan, D, Olland, A, Suto, R.K. | | 登録日 | 2020-04-21 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Antibody-mediated inhibition of GDF15-GFRAL activity reverses cancer cachexia in mice.
Nat Med, 26, 2020
|
|
1SNB
 
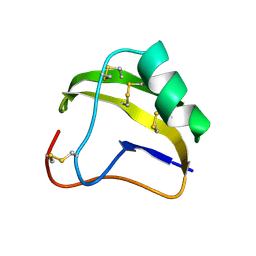 | |
6LYX
 
 | | Crystal structure of oxidized ACHT1 | | 分子名称: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | 著者 | Wang, J.C, Pan, W.M, Cai, W.G, Wang, M.Z, Zhang, M. | | 登録日 | 2020-02-16 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
6LYW
 
 | | Structural insight into the biological functions of Arabidopsis thaliana ACHT1 | | 分子名称: | GLYCEROL, SULFATE ION, Thioredoxin-like 2-1, ... | | 著者 | Wang, J.C, Pan, W.M, Wang, M.Z, Zhang, M. | | 登録日 | 2020-02-16 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insight into the biological functions of Arabidopsis thaliana ACHT1.
Int.J.Biol.Macromol., 158, 2020
|
|
9BIG
 
 | | Stat6 bound to degrader AK-1690 | | 分子名称: | Signal transducer and activator of transcription 6, [(2-{[(2S)-1-{(2S,4S)-4-[(7-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}hept-6-yn-1-yl)oxy]-2-[(2R)-2-phenylmorpholine-4-carbonyl]pyrrolidin-1-yl}-3,3-dimethyl-1-oxobutan-2-yl]carbamoyl}-1-benzothiophen-5-yl)di(fluoro)methyl]phosphonic acid | | 著者 | Mallik, L, Stuckey, J.A. | | 登録日 | 2024-04-23 | | 公開日 | 2024-10-02 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (3.304 Å) | | 主引用文献 | Discovery of AK-1690: A Potent and Highly Selective STAT6 PROTAC Degrader.
J.Med.Chem., 68, 2025
|
|
4DBF
 
 | | Crystal structures of Cg1458 | | 分子名称: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION | | 著者 | Ran, T.T, Xu, D.Q, Wang, W.W, Gao, Y.Y, Wang, M.T. | | 登録日 | 2012-01-15 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
8X5D
 
 | |
5WQE
 
 | |
7FHJ
 
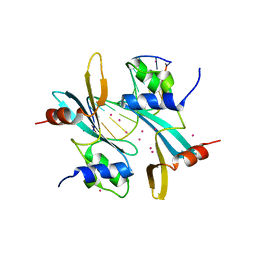 | | Crystal structure of BAZ2A with DNA | | 分子名称: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | 著者 | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | 登録日 | 2021-07-29 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
5WTK
 
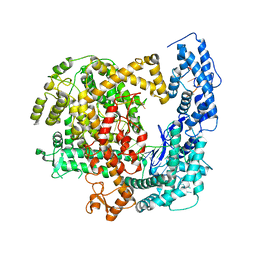 | | Crystal structure of RNP complex | | 分子名称: | CRISPR-associated endoribonuclease C2c2, RNA (58-MER) | | 著者 | Liu, L, Wang, Y. | | 登録日 | 2016-12-13 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.651 Å) | | 主引用文献 | Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities
Cell, 168, 2017
|
|
5WTJ
 
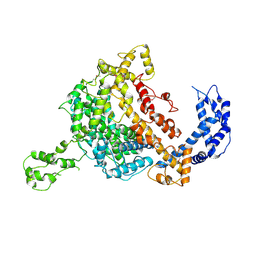 | | Crystal structure of an endonuclease | | 分子名称: | CRISPR-associated endoribonuclease C2c2 | | 著者 | Liu, L, Wang, Y. | | 登録日 | 2016-12-13 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.503 Å) | | 主引用文献 | Two Distant Catalytic Sites Are Responsible for C2c2 RNase Activities
Cell, 168, 2017
|
|
3BZU
 
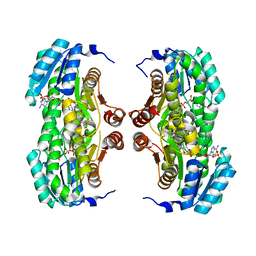 | | Crystal structure of human 11-beta-hydroxysteroid dehydrogenase(HSD1) in complex with NADP and thiazolone inhibitor | | 分子名称: | (5S)-2-{[(1S)-1-(2-fluorophenyl)ethyl]amino}-5-methyl-5-(trifluoromethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Min, X, Sudom, A, Xu, H, Wang, Z, Walker, N.P. | | 登録日 | 2008-01-18 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural characterization and pharmacodynamic effects of an orally active 11beta-hydroxysteroid dehydrogenase type 1 inhibitor.
Chem.Biol.Drug Des., 71, 2008
|
|
5GNF
 
 | |
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | 分子名称: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | 著者 | Zhang, X, Ma, J, Wang, Y, Wang, J. | | 登録日 | 2016-08-07 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
5XPY
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | 分子名称: | ACETATE ION, Fermitin family homolog 2, GLYCEROL | | 著者 | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | 登録日 | 2017-06-05 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPZ
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | 分子名称: | Fermitin family homolog 2, GLYCEROL | | 著者 | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | 登録日 | 2017-06-05 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
