7DK3
 
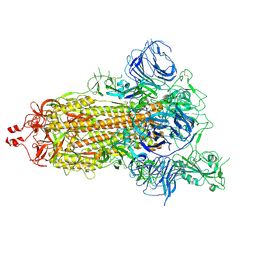 | | SARS-CoV-2 S trimer, S-open | | 分子名称: | Spike glycoprotein | | 著者 | Xu, C, Cong, Y. | | 登録日 | 2020-11-23 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
3SJC
 
 | |
7DF4
 
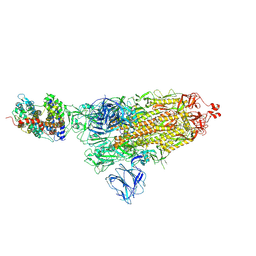 | | SARS-CoV-2 S-ACE2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Cong, X, Yao, C. | | 登録日 | 2020-11-06 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DF3
 
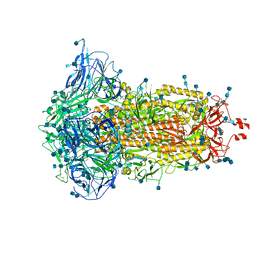 | | SARS-CoV-2 S trimer, S-closed | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cong, X, Yao, C. | | 登録日 | 2020-11-06 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
3SJA
 
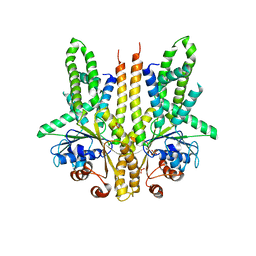 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | 分子名称: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | 著者 | Reitz, S, Wild, K, Sinning, I. | | 登録日 | 2011-06-21 | | 公開日 | 2011-07-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | 著者 | Rao, X, Chai, Y, Wu, Y, Gao, F. | | 登録日 | 2022-06-11 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5Z8F
 
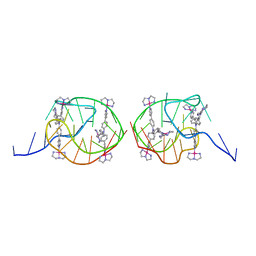 | | Solution structure for the unique dimeric 4:2 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | 分子名称: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | 著者 | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | 登録日 | 2018-01-31 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
8WYP
 
 | |
7E7E
 
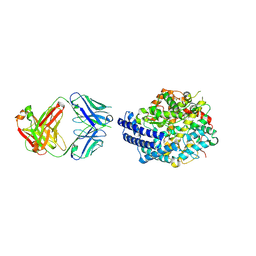 | | The co-crystal structure of ACE2 with Fab | | 分子名称: | Processed angiotensin-converting enzyme 2, ZINC ION, h11B11-Fab | | 著者 | Xiao, J.Y, Zhang, Y. | | 登録日 | 2021-02-26 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7F75
 
 | | Cryo-EM structure of Spx-dependent transcription activation complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Lin, W, Feng, Y, Shi, J. | | 登録日 | 2021-06-28 | | 公開日 | 2021-10-13 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of transcription activation by the global regulator Spx.
Nucleic Acids Res., 49, 2021
|
|
2CCA
 
 | |
5Z80
 
 | | Solution structure for the 1:1 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | 分子名称: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | 著者 | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | 登録日 | 2018-01-30 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
2CCD
 
 | |
6UIF
 
 | |
8EXH
 
 | | Agrobacterium tumefaciens Tpilus | | 分子名称: | (14S,17R)-20-amino-17-hydroxy-11,17-dioxo-12,16,18-trioxa-17lambda~5~-phosphaicosan-14-yl tetradecanoate, Protein virB2 | | 著者 | Beltran, L.C, Egelman, E.H. | | 登録日 | 2022-10-25 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery
Nat Commun, 14, 2023
|
|
6UIK
 
 | |
6UJH
 
 | |
8SK4
 
 | |
8SKH
 
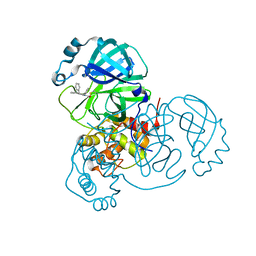 | | Co-structure of SARS-CoV-2 (COVID-19 with covalent pyrazoline based inhibitors | | 分子名称: | 2-chloro-1-[(4R,5R)-3,4,5-triphenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase nsp5 | | 著者 | Knapp, M.S, Ornelas, E. | | 登録日 | 2023-04-19 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.882 Å) | | 主引用文献 | Discovery of Potent Pyrazoline-Based Covalent SARS-CoV-2 Main Protease Inhibitors.
Chembiochem, 24, 2023
|
|
6UJ4
 
 | |
6UJL
 
 | |
6UTF
 
 | |
6VG0
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC ISOCITRATE DEHYDROGENASE (IDH1) R132H MUTANT IN COMPLEX WITH NADPH and AGI-15056 | | 分子名称: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~2~,N~4~-bis[(1R)-1-cyclopropylethyl]-6-[6-(trifluoromethyl)pyridin-2-yl]-1,3,5-triazine-2,4-diamine | | 著者 | Padyana, A, Jin, L. | | 登録日 | 2020-01-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Vorasidenib (AG-881): A First-in-Class, Brain-Penetrant Dual Inhibitor of Mutant IDH1 and 2 for Treatment of Glioma.
Acs Med.Chem.Lett., 11, 2020
|
|
6UTH
 
 | |
6UTJ
 
 | |
