7M6T
 
 | | Crystal structure of SOCS2/ElonginB/ElonginC bound to a non-canonical peptide that enhances phospho-peptide binding | | 分子名称: | Elongin-B, Elongin-C, Non-canonical peptide F3, ... | | 著者 | Kershaw, N.J, Li, K, Linossi, E.M, Nicholson, S.E. | | 登録日 | 2021-03-26 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.194 Å) | | 主引用文献 | Discovery of an exosite on the SOCS2-SH2 domain that enhances SH2 binding to phosphorylated ligands.
Nat Commun, 12, 2021
|
|
6ZTB
 
 | | Crystal Structure of human P-Cadherin EC1_EC2 | | 分子名称: | CALCIUM ION, Cadherin-3, SODIUM ION | | 著者 | Rondeau, J.M, Lehmann, S. | | 登録日 | 2020-07-17 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTF
 
 | |
6ZTR
 
 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 in complex with human P-Cadherin(108-324) | | 分子名称: | CALCIUM ION, CQY684 Fab heavy-chain, CQY684 Fab light-chain, ... | | 著者 | Rondeau, J.M, Lehmann, S. | | 登録日 | 2020-07-20 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
8ELO
 
 | |
8ELQ
 
 | |
8ELP
 
 | |
7T0O
 
 | |
7T0R
 
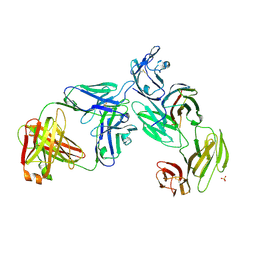 | | Crystal structure of the anti-CD4 adnectin 6940_B01 as a complex with the extracellular domains of CD4 and ibalizumab fAb | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adnectin 6940_B01, Ibalizumab Heavy Chain, ... | | 著者 | Williams, S.P, Concha, N.O, Wensel, D.L, Hong, X. | | 登録日 | 2021-11-30 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.65 Å) | | 主引用文献 | Novel Bent Conformation of CD4 Induced by HIV-1 Inhibitor Indirectly Prevents Productive Viral Attachment.
J.Mol.Biol., 434, 2021
|
|
5IV5
 
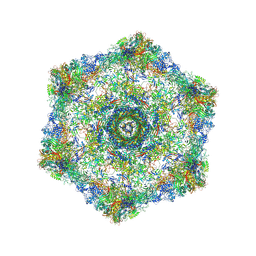 | | Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex | | 分子名称: | Baseplate hub protein gp27, Baseplate tail-tube protein gp48, Baseplate tail-tube protein gp54, ... | | 著者 | Taylor, N.M.I, Guerrero-Ferreira, R.C, Goldie, K.N, Stahlberg, H, Leiman, P.G. | | 登録日 | 2016-03-19 | | 公開日 | 2016-05-18 | | 最終更新日 | 2018-02-07 | | 実験手法 | ELECTRON MICROSCOPY (4.11 Å) | | 主引用文献 | Structure of the T4 baseplate and its function in triggering sheath contraction.
Nature, 533, 2016
|
|
5IV7
 
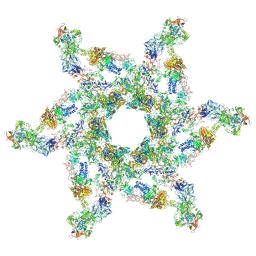 | | Cryo-electron microscopy structure of the star-shaped, hubless post-attachment T4 baseplate | | 分子名称: | Baseplate wedge protein gp10, Baseplate wedge protein gp11, Baseplate wedge protein gp25, ... | | 著者 | Taylor, N.M.I, Guerrero-Ferreira, R.C, Goldie, K.N, Stahlberg, H, Leiman, P.G. | | 登録日 | 2016-03-19 | | 公開日 | 2016-05-18 | | 最終更新日 | 2018-02-07 | | 実験手法 | ELECTRON MICROSCOPY (6.77 Å) | | 主引用文献 | Structure of the T4 baseplate and its function in triggering sheath contraction.
Nature, 533, 2016
|
|
5JDT
 
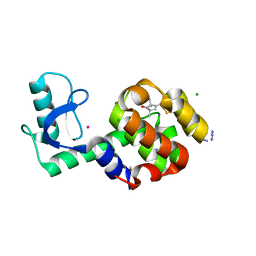 | | Structure of Spin-labelled T4 lysozyme mutant L118C-R1 at 100K | | 分子名称: | AZIDE ION, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | 登録日 | 2016-04-17 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Tracking Transient Conformational States of T4 Lysozyme at Room Temperature Combining X-ray Crystallography and Site-Directed Spin Labeling.
J.Am.Chem.Soc., 138, 2016
|
|
8D6H
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V after UV irradiation | | 分子名称: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, ACETATE ION, ... | | 著者 | Bingham, C.R, Geiger, J.H, Borhan, B. | | 登録日 | 2022-06-06 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
8D6N
 
 | | Q108K:K40L:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V | | 分子名称: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, ACETATE ION, ... | | 著者 | Bingham, C.R, Geiger, J.H, Borhan, B. | | 登録日 | 2022-06-06 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
8DB2
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F mutant of hCRBPII bound to synthetic fluorophore CM1V | | 分子名称: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, Retinol-binding protein 2 | | 著者 | Bingham, C.R, Geiger, J.H, Borhan, B, Staples, R. | | 登録日 | 2022-06-14 | | 公開日 | 2023-02-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
8D6L
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V | | 分子名称: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, GLYCEROL, ... | | 著者 | Bingham, C.R, Geiger, J.H, Borhan, B. | | 登録日 | 2022-06-06 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
1CGC
 
 | |
1BD1
 
 | |
1D26
 
 | |
7OOJ
 
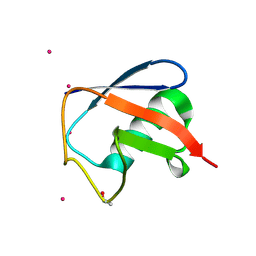 | | Structure of D-Thr53 Ubiquitin | | 分子名称: | CADMIUM ION, Ubiquitin | | 著者 | Becker, S. | | 登録日 | 2021-05-27 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A litmus test for classifying recognition mechanisms of transiently binding proteins.
Nat Commun, 13, 2022
|
|
6XF4
 
 | | Crystal structure of STING REF variant in complex with E7766 | | 分子名称: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), 1,2-ETHANEDIOL, Stimulator of interferon genes protein | | 著者 | Chen, Y, Wang, J.Y, Kim, D.-S. | | 登録日 | 2020-06-15 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
6XF3
 
 | | Crystal structure of STING in complex with E7766 | | 分子名称: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), Stimulator of interferon genes protein | | 著者 | Chen, Y, Wang, J.Y, Kim, D.-S. | | 登録日 | 2020-06-15 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
6U3Z
 
 | | Structure of VD20_5A4 Fab | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Dionne, G, Shapiro, L. | | 登録日 | 2019-08-22 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.99002314 Å) | | 主引用文献 | Extensive dissemination and intraclonal maturation of HIV Env vaccine-induced B cell responses.
J.Exp.Med., 217, 2020
|
|
6U9U
 
 | | Structure of GM9_TH8seq732127 FAB | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, GM9_TH8seq732127 FAB heavy chain, ... | | 著者 | Singh, S, Liban, T.J, Pancera, M. | | 登録日 | 2019-09-09 | | 公開日 | 2019-11-06 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Extensive dissemination and intraclonal maturation of HIV Env vaccine-induced B cell responses.
J.Exp.Med., 217, 2020
|
|
5EYM
 
 | | MEK1 IN COMPLEX WITH BI 847325 | | 分子名称: | 3-[(3~{Z})-3-[[[4-[(dimethylamino)methyl]phenyl]amino]-phenyl-methylidene]-2-oxidanylidene-1~{H}-indol-6-yl]-~{N}-ethyl-prop-2-ynamide, CALCIUM ION, Dual specificity mitogen-activated protein kinase kinase 1 | | 著者 | Bader, G, Reiser, U, Zahn, S.K, Treu, M. | | 登録日 | 2015-11-25 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases.
Mol.Cancer Ther., 15, 2016
|
|
