1UDE
 
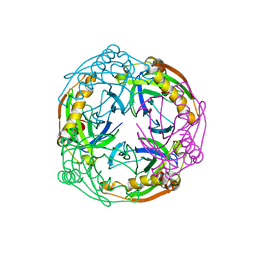 | | Crystal structure of the Inorganic pyrophosphatase from the hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | 分子名称: | Inorganic pyrophosphatase | | 著者 | Liu, B, Gao, R, Zhou, W, Bartlam, M, Rao, Z. | | 登録日 | 2003-04-29 | | 公開日 | 2004-01-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Crystal structure of the hyperthermophilic inorganic pyrophosphatase from the archaeon Pyrococcus horikoshii.
Biophys.J., 86, 2004
|
|
7EW4
 
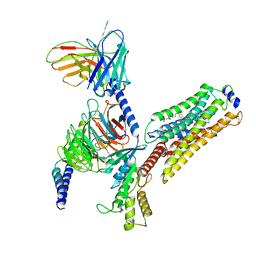 | | Cryo-EM structure of CYM-5541-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW2
 
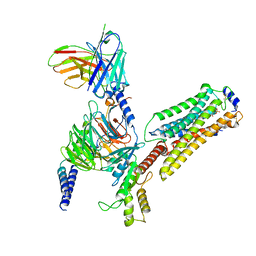 | | Cryo-EM structure of pFTY720-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | 分子名称: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
7EW3
 
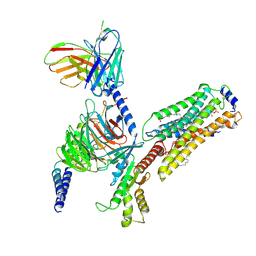 | | Cryo-EM structure of S1P-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | 分子名称: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
1RKN
 
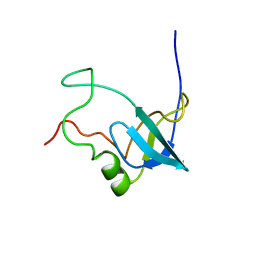 | | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | | 分子名称: | Thermonuclease | | 著者 | Liu, D.S, Feng, Y.G, Ye, K.Q, Shan, L, Wang, J.F. | | 登録日 | 2003-11-22 | | 公開日 | 2004-12-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
7D1M
 
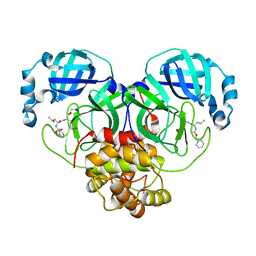 | | CRYSTAL STRUCTURE OF THE SARS-CoV-2 MAIN PROTEASE COMPLEXED WITH GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fu, L.F, Gilski, M, Shabalin, I, Gao, G.F, Qi, J.X. | | 登録日 | 2020-09-14 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
5E9A
 
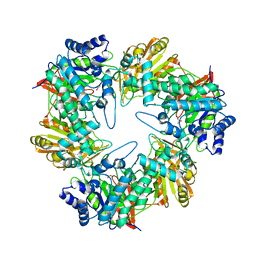 | |
7CCF
 
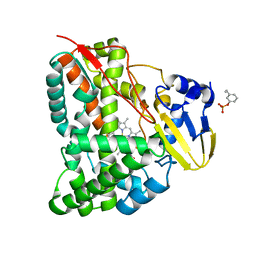 | | Mechanism insights on steroselective oxidation of phosphorylated ethylphenols with cytochrome P450 CreJ | | 分子名称: | (3-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Dong, S, Du, L, Li, S.Y, Feng, Y.G. | | 登録日 | 2020-06-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis for Selective Oxidation of Phosphorylated Ethylphenols by Cytochrome P450 Monooxygenase CreJ.
Appl.Environ.Microbiol., 87, 2021
|
|
4OIR
 
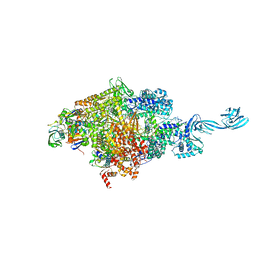 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | 分子名称: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.105 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | 分子名称: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.624 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
6JOM
 
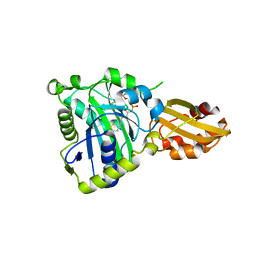 | |
8HN6
 
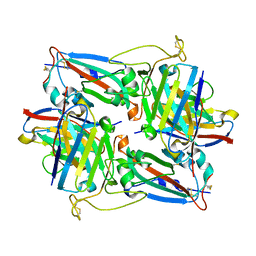 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | 分子名称: | Heavy chain of monoclonal antibody 3G10, Light chain of monoclonal antibody 3G10, Spike protein S1 | | 著者 | Qi, J, Chen, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
8HN7
 
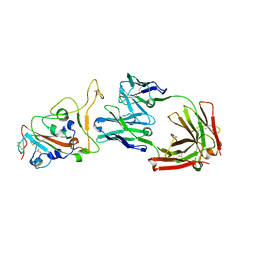 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 3C11, Light chain of monoclonal antibody 3C11, ... | | 著者 | Qi, J, Chen, Y. | | 登録日 | 2022-12-07 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-06-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
7EU0
 
 | | The cryo-EM structure of A. thaliana Pol IV-RDR2 backtracked complex | | 分子名称: | DNA (33-MER), DNA (5'-D(*CP*TP*GP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*GP*AP*GP*TP*G)-3'), DNA-directed RNA polymerase IV subunit 1, ... | | 著者 | Fang, C.L, Wu, X.X, Huang, K, Zhang, Y. | | 登録日 | 2021-05-15 | | 公開日 | 2021-12-29 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Science, 374, 2021
|
|
7EU1
 
 | | The cryo-EM structure of A. thaliana Pol IV-RDR2 holoenzyme | | 分子名称: | DNA-directed RNA polymerase IV subunit 1, DNA-directed RNA polymerases II and IV subunit 5A, DNA-directed RNA polymerases II, ... | | 著者 | Fang, C.L, Wu, X.X, Huang, K, Zhang, Y. | | 登録日 | 2021-05-15 | | 公開日 | 2021-12-29 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Science, 374, 2021
|
|
4OIN
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | 分子名称: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIO
 
 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | 分子名称: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4PNJ
 
 | | Recombinant Sperm Whale P6 Myoglobin Solved with Single Pulse Free Electron Laser Data | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | 著者 | Cohen, A, Gonzalez, A, Lam, W, Lyubimov, A, Sauter, N, Tsai, Y, Uervirojnangkoorn, M, Brunger, A, Soltis, M. | | 登録日 | 2014-05-23 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Goniometer-based femtosecond crystallography with X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6K1T
 
 | |
8XTG
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and DMMPA | | 分子名称: | O-desmethyl mycophenolic acid, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | 登録日 | 2024-01-10 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
8XTF
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and FDHMP-3C | | 分子名称: | 4-farnesyl-3,5-dihydroxy-6-methylphthalide-3C, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | 登録日 | 2024-01-10 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
8XTE
 
 | | Crystal structure of methyltransferase MpaG' in complex with SAH and FDHMP | | 分子名称: | 4-farnesyl-3,5-dihydroxy-6-methylphthalide, O-methyltransferase mpaG', S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | You, C, Pan, Y.J, Li, S.Y, Feng, Y.G. | | 登録日 | 2024-01-10 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural basis for substrate flexibility of the O-methyltransferase MpaG' involved in mycophenolic acid biosynthesis.
Protein Sci., 33, 2024
|
|
4OIP
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | 分子名称: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | 著者 | Zhang, Y, Ebright, R.H, Arnold, E. | | 登録日 | 2014-01-20 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
7F0R
 
 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | 分子名称: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | He, D.W, You, L.L, Zhang, Y. | | 登録日 | 2021-06-06 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (5.8 Å) | | 主引用文献 | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7WDI
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87K in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | 分子名称: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | 著者 | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | 登録日 | 2021-12-21 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
