5YCO
 
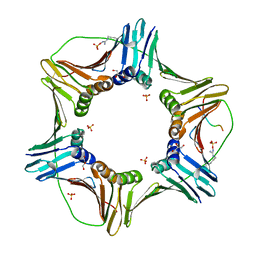 | | Complex structure of PCNA with UHRF2 | | 分子名称: | E3 ubiquitin-protein ligase UHRF2, GLYCEROL, Proliferating cell nuclear antigen, ... | | 著者 | Wu, M, Chen, W, Hang, T, Wang, C, Zhang, X, Zang, J. | | 登録日 | 2017-09-07 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
2HMY
 
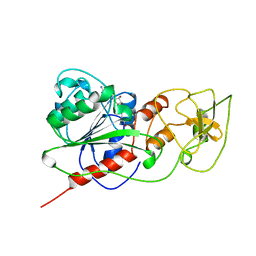 | | BINARY COMPLEX OF HHAI METHYLTRANSFERASE WITH ADOMET FORMED IN THE PRESENCE OF A SHORT NONPSECIFIC DNA OLIGONUCLEOTIDE | | 分子名称: | PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), S-ADENOSYLMETHIONINE | | 著者 | O'Gara, M, Zhang, X, Roberts, R.J, Cheng, X. | | 登録日 | 1999-02-08 | | 公開日 | 1999-03-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structure of a binary complex of HhaI methyltransferase with S-adenosyl-L-methionine formed in the presence of a short non-specific DNA oligonucleotide.
J.Mol.Biol., 287, 1999
|
|
7NTS
 
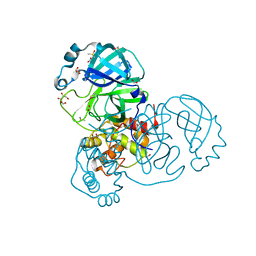 | | Crystal structure of the SARS-CoV-2 Main Protease with oxidized C145 | | 分子名称: | DIMETHYL SULFOXIDE, FORMIC ACID, GLYCEROL, ... | | 著者 | Dupre, E, Villeret, V, Hanoulle, X. | | 登録日 | 2021-03-10 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.477 Å) | | 主引用文献 | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5COA
 
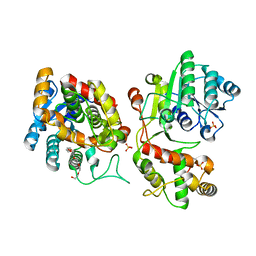 | | Crystal structure of iridoid synthase at 2.2-angstrom resolution | | 分子名称: | HEXAETHYLENE GLYCOL, Iridoid synthase, SULFATE ION | | 著者 | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
7NTQ
 
 | | Crystal structure of the SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | 著者 | Dupre, E, Villeret, V, Hanoulle, X. | | 登録日 | 2021-03-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.495 Å) | | 主引用文献 | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
7NTW
 
 | |
7NTT
 
 | |
5COB
 
 | | Crystal structure of iridoid synthase in complex with NADP+ and 8-oxogeranial at 2.65-angstrom resolution | | 分子名称: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Qin, L, Zhu, Y, Ding, Z, Zhang, X, Ye, S, Zhang, R. | | 登録日 | 2015-07-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of iridoid synthase in complex with NADP(+)/8-oxogeranial reveals the structural basis of its substrate specificity.
J.Struct.Biol., 194, 2016
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
3J6C
 
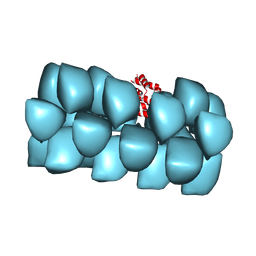 | | Cryo-EM structure of MAVS CARD filament | | 分子名称: | Mitochondrial antiviral-signaling protein | | 著者 | Xu, H, He, X, Zheng, H, Huang, L.J, Hou, F, Yu, Z, de la Cruz, M.J, Borkowski, B, Zhang, X, Chen, Z.J, Jiang, Q.-X. | | 登録日 | 2014-02-04 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.6 Å) | | 主引用文献 | Structural basis for the prion-like MAVS filaments in antiviral innate immunity.
Elife, 3, 2014
|
|
3KV5
 
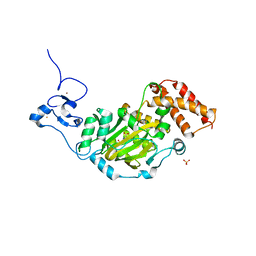 | | Structure of KIAA1718, human Jumonji demethylase, in complex with N-oxalylglycine | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV9
 
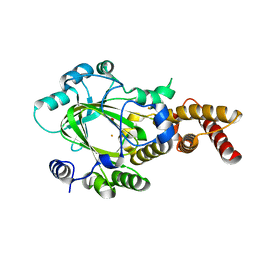 | | Structure of KIAA1718 Jumonji domain | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5CMS
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, SULFATE ION | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-07-17 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
3KVB
 
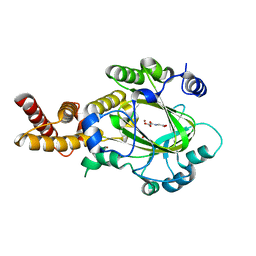 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | 分子名称: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVA
 
 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV4
 
 | | Structure of PHF8 in complex with histone H3 | | 分子名称: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5CB3
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase | | 著者 | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | 登録日 | 2015-06-30 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
3KV6
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2R5W
 
 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | 著者 | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | 登録日 | 2007-09-04 | | 公開日 | 2008-03-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
1P9E
 
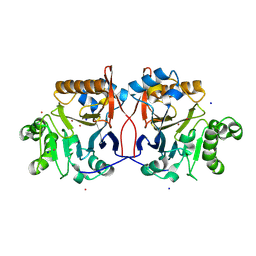 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | 分子名称: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | 著者 | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | 登録日 | 2003-05-11 | | 公開日 | 2004-05-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3
To be Published
|
|
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | 分子名称: | NLR family CARD domain-containing protein 4 | | 著者 | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | 登録日 | 2015-09-05 | | 公開日 | 2015-10-21 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
8BQN
 
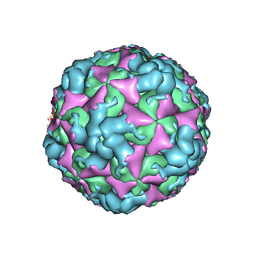 | |
8PII
 
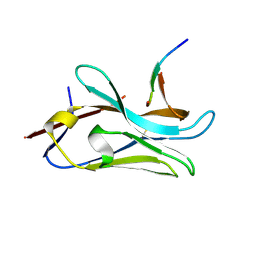 | | VHH Z70 mutant 3 in interaction with PHF6 Tau peptide | | 分子名称: | Microtubule-associated protein tau, VHH Z70 Mutant 3 | | 著者 | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | 登録日 | 2023-06-21 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
5EWU
 
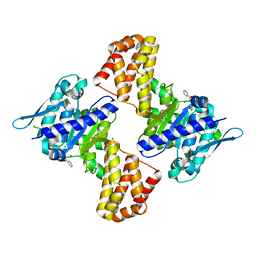 | | Crystal structure of the Arabidopsis thaliana C-terminal Chlh at 1.25A | | 分子名称: | BENZOIC ACID, MAGNESIUM ION, Magnesium-chelatase subunit ChlH, ... | | 著者 | Chen, Z, Zhang, X, Liu, Y, Jiang, L. | | 登録日 | 2015-11-21 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal structure of the Arabidopsis thaliana C-terminal Chlh at 1.25A
To Be Published
|
|
5ABH
 
 | | Structure of GH84 with ligand | | 分子名称: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | 著者 | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | 登録日 | 2015-08-05 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
