8EM4
 
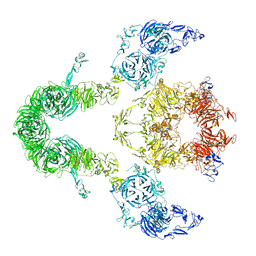 | | Cryo-EM structure of LRP2 at pH 7.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Beenken, A, Cerutti, G, Brasch, J, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | 登録日 | 2022-09-26 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
6Q89
 
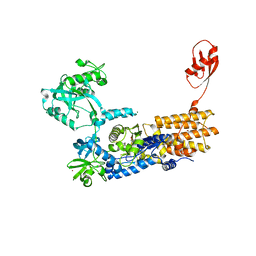 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with the Intermediate Analog 5'-O-(N-(L-Leucyl)-Sulfamoyl)Adenosine | | 分子名称: | 1,2-ETHANEDIOL, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | 著者 | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | 登録日 | 2018-12-14 | | 公開日 | 2019-04-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Comparative analysis of pyrimidine substituted aminoacyl-sulfamoyl nucleosides as potential inhibitors targeting class I aminoacyl-tRNA synthetases.
Eur.J.Med.Chem., 173, 2019
|
|
5ZNS
 
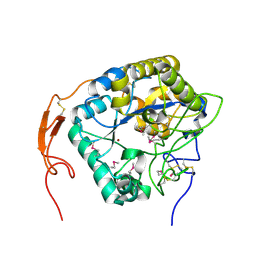 | | Insect chitin deacetylase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ZINC ION, ... | | 著者 | Liu, L, Zhou, Y, Yang, Q. | | 登録日 | 2018-04-10 | | 公開日 | 2019-02-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Structural and biochemical insights into the catalytic mechanisms of two insect chitin deacetylases of the carbohydrate esterase 4 family.
J. Biol. Chem., 294, 2019
|
|
6Q8V
 
 | |
4A9C
 
 | | Crystal structure of human SHIP2 in complex with biphenyl 2,3',4,5',6- pentakisphosphate | | 分子名称: | BIPHENYL 2,3',4,5',6-PENTAKISPHOSPHATE, PHOSPHATIDYLINOSITOL-3,4,5-TRISPHOSPHATE 5-PHOSPHATASE 2 | | 著者 | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Graslund, S, Karlberg, T, Mills, S.J, Moche, M, Nyman, T, Persson, C, Potter, B.V.L, Schuler, H, Thorsell, A.G, Weigelt, J, Nordlund, P. | | 登録日 | 2011-11-25 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Synthetic Polyphosphoinositide Headgroup Surrogate in Complex with Ship2 Provides a Rationale for Drug Discovery.
Acs Chem.Biol., 7, 2012
|
|
6Q8Y
 
 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2018-12-16 | | 公開日 | 2019-03-13 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7MEQ
 
 | | Crystal structure of human TMPRSS2 in complex with Nafamostat | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Transmembrane protease serine 2, ... | | 著者 | Fraser, B, Beldar, S, Hutchinson, A, Li, Y, Seitova, A, Edwards, A.M, Benard, F, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2021-04-07 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation.
Nat.Chem.Biol., 18, 2022
|
|
4AA7
 
 | | E.coli GlmU in complex with an antibacterial inhibitor | | 分子名称: | BIFUNCTIONAL PROTEIN GLMU, N-(2,4-dimethoxy-5-{[(2R)-2-methyl-2,3-dihydro-1H-indol-1-yl]sulfonyl}phenyl)acetamide, SULFATE ION | | 著者 | Otterbein, L, Breed, J, Ogg, D.J. | | 登録日 | 2011-11-30 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6QDY
 
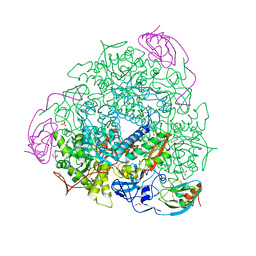 | | The crystal structure of Sporosarcina pasteurii urease in complex with its substrate urea | | 分子名称: | 1,2-ETHANEDIOL, FLUORIDE ION, NICKEL (II) ION, ... | | 著者 | Mazzei, L, Cianci, M, Benini, S, Ciurli, S. | | 登録日 | 2019-01-03 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.416 Å) | | 主引用文献 | The Structure of the Elusive Urease-Urea Complex Unveils the Mechanism of a Paradigmatic Nickel-Dependent Enzyme.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4AAW
 
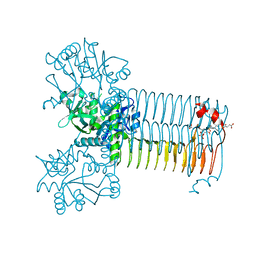 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | 分子名称: | 4-{[1-(2-{[({5-[(3-carboxypropanoyl)amino]-2,4-dimethoxyphenyl}sulfonyl)amino]methyl}phenyl)piperidin-4-yl]methoxy}-4-oxobutanoic acid, BIFUNCTIONAL PROTEIN GLMU, SULFATE ION | | 著者 | Otterbein, L, Breed, J, Ogg, D.J. | | 登録日 | 2011-12-05 | | 公開日 | 2012-02-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6QK9
 
 | |
6QLE
 
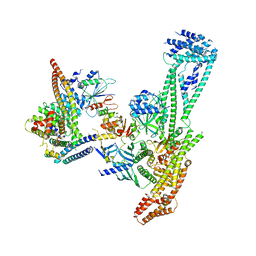 | | Structure of inner kinetochore CCAN complex | | 分子名称: | Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3,Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3, Central kinetochore subunit MCM16,Central kinetochore subunit MCM16,Inner kinetochore subunit MCM16,Mcm16p, Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1, ... | | 著者 | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | 登録日 | 2019-01-31 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.55 Å) | | 主引用文献 | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
4A29
 
 | | Structure of the engineered retro-aldolase RA95.0 | | 分子名称: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, D-MALATE, ENGINEERED RETRO-ALDOL ENZYME RA95.0 | | 著者 | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | 登録日 | 2011-09-23 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A2R
 
 | | Structure of the engineered retro-aldolase RA95.5-5 | | 分子名称: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | 著者 | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | 登録日 | 2011-09-28 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.302 Å) | | 主引用文献 | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A3R
 
 | | Crystal structure of Enolase from Bacillus subtilis. | | 分子名称: | CITRIC ACID, ENOLASE, SODIUM ION | | 著者 | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A.S, Harwood, C.R, Lewis, R.J. | | 登録日 | 2011-10-04 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Dissection of the Network of Interactions that Links RNA Processing with Glycolysis in the Bacillus Subtilis Degradosome.
J.Mol.Biol., 416, 2012
|
|
6PT0
 
 | | Cryo-EM structure of human cannabinoid receptor 2-Gi protein in complex with agonist WIN 55,212-2 | | 分子名称: | CHOLESTEROL, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Xu, T.H, Xing, C, Zhuang, Y, Feng, Z, Zhou, X.E, Chen, M, Wang, L, Meng, X, Xue, Y, Wang, J, Liu, H, McGuire, T, Zhao, G, Melcher, K, Zhang, C, Xu, H.E, Xie, X.Q. | | 登録日 | 2019-07-14 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-04 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM Structure of the Human Cannabinoid Receptor CB2-GiSignaling Complex.
Cell, 180, 2020
|
|
6QIV
 
 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL4 compound | | 分子名称: | CMBL4, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | 著者 | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | 登録日 | 2019-01-21 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
4A3S
 
 | | Crystal structure of PFK from Bacillus subtilis | | 分子名称: | 6-PHOSPHOFRUCTOKINASE | | 著者 | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A.S, Harwood, C.R, Lewis, R.J. | | 登録日 | 2011-10-04 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Dissection of the Network of Interactions that Links RNA Processing with Glycolysis in the Bacillus Subtilis Degradosome.
J.Mol.Biol., 416, 2012
|
|
4AC3
 
 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | 分子名称: | BIFUNCTIONAL PROTEIN GLMU, N-{2,4-DIMETHOXY-5-[(2-PIPERIDIN-1-YLBENZYL)sulfamoyl]phenyl}acetamide, SULFATE ION | | 著者 | Otterbein, L, Breed, J, Ogg, D.J. | | 登録日 | 2011-12-12 | | 公開日 | 2012-02-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7WJY
 
 | | Omicron spike trimer with 6m6 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-08 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJZ
 
 | | Omicron Spike bitrimer with 6m6 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | 登録日 | 2022-01-08 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
4A1F
 
 | | Crystal structure of C-terminal domain of Helicobacter pylori DnaB Helicase | | 分子名称: | CITRATE ANION, REPLICATIVE DNA HELICASE | | 著者 | Stelter, M, Kapp, U, Timmins, J, Terradot, L. | | 登録日 | 2011-09-14 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Architecture of a Dodecameric Bacterial Replicative Helicase.
Structure, 20, 2012
|
|
8EUS
 
 | |
4AJM
 
 | | Development of a plate-based optical biosensor methodology to identify PDE10 fragment inhibitors | | 分子名称: | 3-AMINO-6-FLUORO-2-[4-(2-METHYLPYRIDIN-4-YL)PHENYL]-N-(METHYLSULFONYL)QUINOLINE-4-CARBOXAMIDE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | 著者 | Geschwindner, S, Johansson, P, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Albert, J.S. | | 登録日 | 2012-02-16 | | 公開日 | 2013-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Development of a Plate-Based Optical Biosensor Methodology to Identify Pde10 Fragment Inhibitors
To be Published
|
|
4AK6
 
 | | BpGH117_H302E mutant glycoside hydrolase | | 分子名称: | ANHYDRO-ALPHA-L-GALACTOSIDASE, MAGNESIUM ION | | 著者 | Hehemann, J.H, Smyth, L, Yadav, A, Vocadlo, D.J, Boraston, A.B. | | 登録日 | 2012-02-21 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Analysis of Keystone Enzyme in Agar Hydrolysis Provides Insight Into the Degradation (of a Polysaccharide from) Red Seaweeds.
J.Biol.Chem., 287, 2012
|
|
