2KU7
 
 | |
4QBR
 
 | |
2LA5
 
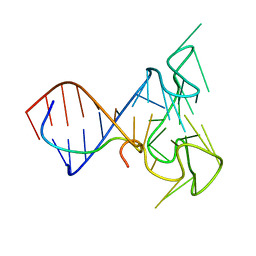 | | RNA Duplex-Quadruplex Junction Complex with FMRP RGG peptide | | 分子名称: | Fragile X mental retardation 1 protein, RNA (36-MER) | | 著者 | Phan, A, Kuryavyi, V, Darnell, J, Serganov, A, Majumdar, A, Ilin, S, Darnell, R, Patel, D. | | 登録日 | 2011-03-03 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4TN7
 
 | | Crystal structure of mouse KDM2A-H3K36ME-NO complex | | 分子名称: | FE (III) ION, Lysine-specific demethylase 2A, NITRIC OXIDE, ... | | 著者 | Cheng, Z. | | 登録日 | 2014-06-03 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36.
Genes Dev., 28, 2014
|
|
4N76
 
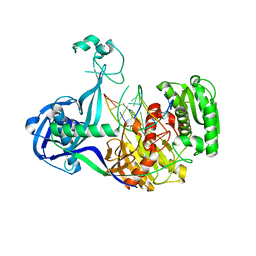 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and cleaved target DNA with Mn2+ | | 分子名称: | 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-15 | | 公開日 | 2014-01-15 | | 最終更新日 | 2025-02-12 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N41
 
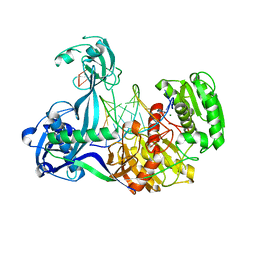 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 15-mer target DNA | | 分子名称: | 5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*GP*CP*CP*TP*CP*G)-3', 5'-D(P*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*T*GP*TP*AP*TP*AP*GP*T)-3', ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-08 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.248 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NCB
 
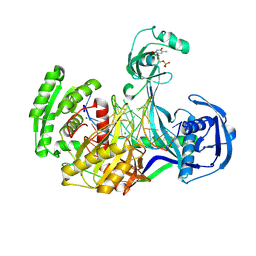 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 19-mer target DNA with Mg2+ | | 分子名称: | 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*C)-3', 5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-24 | | 公開日 | 2014-01-15 | | 最終更新日 | 2025-02-12 | | 実験手法 | X-RAY DIFFRACTION (2.189 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NCA
 
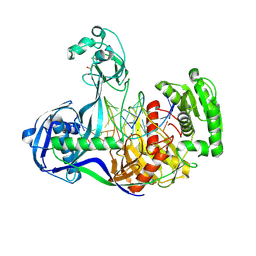 | | Structure of Thermus thermophilus Argonaute bound to guide DNA 19-mer and target DNA in the presence of Mg2+ | | 分子名称: | 5'-D(*AP*CP*AP*AP*CP*C)-3', 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-24 | | 公開日 | 2014-01-15 | | 最終更新日 | 2025-02-12 | | 実験手法 | X-RAY DIFFRACTION (2.489 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N47
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 12-mer target DNA | | 分子名称: | 5'-D(*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-08 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.823 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5ZUU
 
 | | Crystal structure of AtCPSF30 YTH domain in complex with 10mer m6A-modified RNA | | 分子名称: | 30-kDa cleavage and polyadenylation specificity factor 30, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | 著者 | Wu, B, Nie, H, Li, S, Patel, D.J. | | 登録日 | 2018-05-08 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis.
Mol Plant, 2021
|
|
4GIX
 
 | | Crystal structure of human GLTP bound with 12:0 disulfatide | | 分子名称: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | 著者 | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | 登録日 | 2012-08-09 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHS
 
 | | Crystal structure of human GLTP bound with 12:0 disulfatide (orthorombic form; two subunits in asymmetric unit) | | 分子名称: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | 著者 | Samygina, V.R, Popov, A.N, Goni-de-Cerio, F, Cabo-Bilbao, A, Malinina, L. | | 登録日 | 2012-08-08 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHP
 
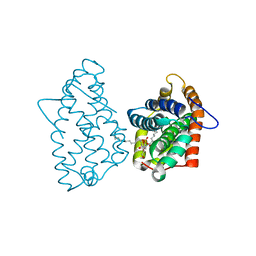 | | Crystal Structure of D48V||A47D mutant of Human GLTP bound with 12:0 monosulfatide | | 分子名称: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | 著者 | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | 登録日 | 2012-08-08 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GJQ
 
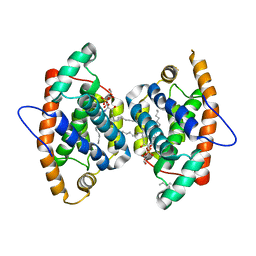 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form;two subunits in asymmetric unit) | | 分子名称: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein, HEXANE | | 著者 | Cabo-Bilbao, A, Goni-de-Cerio, F, Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | 登録日 | 2012-08-10 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GH0
 
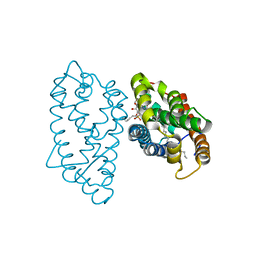 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 monosulfatide | | 分子名称: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE | | 著者 | Samygina, V.R, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | 登録日 | 2012-08-07 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GVT
 
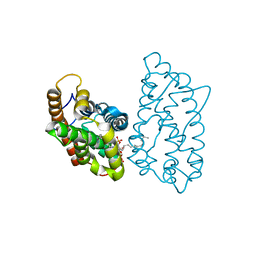 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide (hexagonal form) | | 分子名称: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | 著者 | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | 登録日 | 2012-08-31 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXG
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form; four subunits in asymmetric unit) | | 分子名称: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | 著者 | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | 登録日 | 2012-09-04 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H2Z
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide | | 分子名称: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE, ... | | 著者 | Samygina, V.R, Ochoa-Lizarralde, B, Malinina, L. | | 登録日 | 2012-09-13 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXD
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 disulfatide | | 分子名称: | Glycolipid transfer protein, N-{(2S,3R,4E)-1-[(3,6-di-O-sulfo-beta-D-galactopyranosyl)oxy]-3-hydroxyoctadec-4-en-2-yl}dodecanamide | | 著者 | Samygina, V.R, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | 登録日 | 2012-09-04 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8C58
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-hydroxycytosine and 5-methylcytosine containing dsDNA | | 分子名称: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5OC)P*GP*CP*TP*GP*AP*A)-3'), ... | | 著者 | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C59
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-bromocytosine (converted to 5mC) and 5-methylcytosine containing dsDNA | | 分子名称: | CARBONATE ION, CITRIC ACID, Cytosine-specific methyltransferase, ... | | 著者 | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C57
 
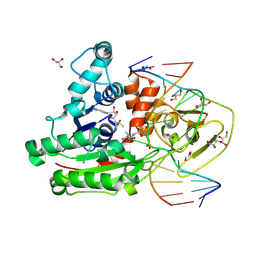 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5,6-dihydro-5-azacytosine (converted to 5m-dhaC) and 5-methylcytosine containing dsDNA | | 分子名称: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5MA)P*GP*CP*TP*GP*AP*A)-3'), ... | | 著者 | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C56
 
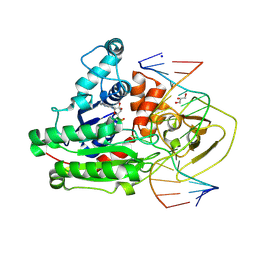 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 2'-deoxy-5-methylzebularine (5mZ) and 5-methylcytosine containing dsDNA | | 分子名称: | Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5PY)P*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*TP*G)-3'), ... | | 著者 | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
4KSA
 
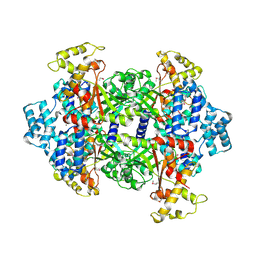 | | Crystal Structure of Malonyl-CoA decarboxylase from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR127 | | 分子名称: | MAGNESIUM ION, Malonyl-CoA decarboxylase | | 著者 | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-05-17 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
9C3G
 
 | |
