5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | 著者 | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | 登録日 | 2018-04-12 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (3.29 Å) | | 主引用文献 | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2P05
 
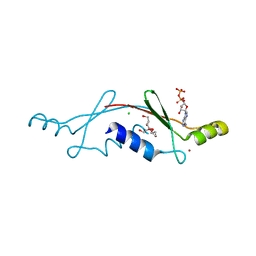 | | Structural Insights into the Evolution of a Non-Biological Protein | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | 著者 | Smith, M, Rosenow, M, Wang, M, Allen, J.P, Szostak, J.W, Chaput, J.C. | | 登録日 | 2007-02-28 | | 公開日 | 2007-06-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the evolution of a non-biological protein: importance of surface residues in protein fold optimization.
PLoS ONE, 2, 2007
|
|
3UYI
 
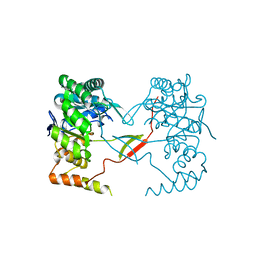 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | 分子名称: | Perakine reductase | | 著者 | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | 登録日 | 2011-12-06 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.313 Å) | | 主引用文献 | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3V0T
 
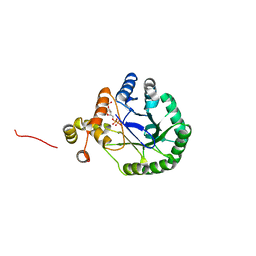 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | 分子名称: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine Reductase | | 著者 | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | 登録日 | 2011-12-08 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.333 Å) | | 主引用文献 | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3UA3
 
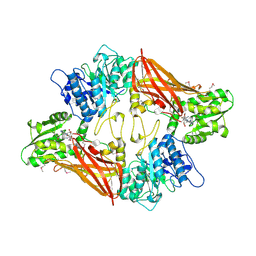 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 in complex with SAH | | 分子名称: | Protein arginine N-methyltransferase 5, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | 登録日 | 2011-10-20 | | 公開日 | 2011-12-14 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3VTQ
 
 | |
3UA4
 
 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 | | 分子名称: | GLYCEROL, Protein arginine N-methyltransferase 5 | | 著者 | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | 登録日 | 2011-10-21 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.005 Å) | | 主引用文献 | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3VGX
 
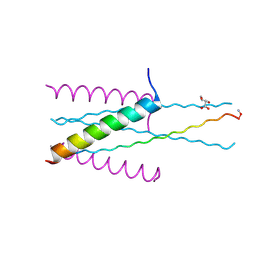 | | Structure of gp41 T21/Cp621-652 | | 分子名称: | ACETIC ACID, Envelope glycoprotein gp160, GLYCEROL | | 著者 | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | 登録日 | 2011-08-22 | | 公開日 | 2012-04-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Discovery of critical residues for viral entry and inhibition through structural Insight of HIV-1 fusion inhibitor CP621-652.
J.Biol.Chem., 287, 2012
|
|
6LW5
 
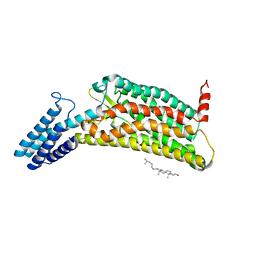 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | 分子名称: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | 著者 | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2020-02-07 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
1KV0
 
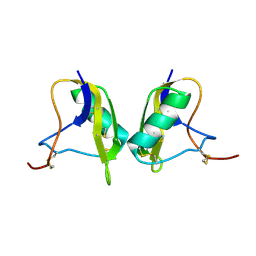 | | Cis/trans Isomerization of Non-prolyl Peptide Bond Observed in Crystal Structure of an Scorpion Toxin | | 分子名称: | Alpha-like toxin BmK-M7 | | 著者 | Guan, R.J, He, X.L, Wang, M, Xiang, Y, Wang, D.C. | | 登録日 | 2002-01-23 | | 公開日 | 2003-09-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural mechanism governing cis and trans isomeric states and an intramolecular switch for cis/trans isomerization of a non-proline peptide bond observed in crystal structures of scorpion toxins.
J.Mol.Biol., 341, 2004
|
|
2MN4
 
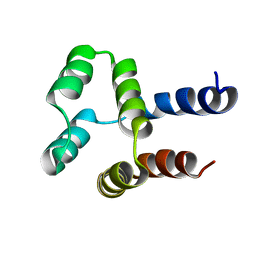 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | 分子名称: | Computational designed protein based on structure template 1cy5 | | 著者 | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | 登録日 | 2014-03-28 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MLB
 
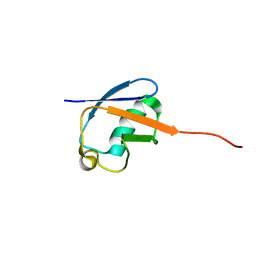 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | 分子名称: | redesigned ubiquitin | | 著者 | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | 登録日 | 2014-02-21 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
6JN2
 
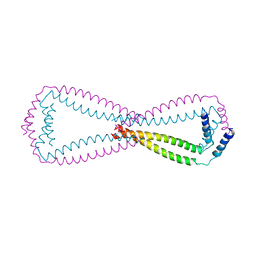 | | Crystal structure of the coiled-coil domains of human DOT1L in complex with AF10 | | 分子名称: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10 | | 著者 | Song, X, Wang, M, Yang, N, Xu, R.M. | | 登録日 | 2019-03-13 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | A higher-order configuration of the heterodimeric DOT1L-AF10 coiled-coil domains potentiates their leukemogenenic activity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8K2B
 
 | | Cryo-EM structure of the human 39S mitoribosome with Tigecycline | | 分子名称: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | 著者 | Li, X, Wang, M, Cheng, J. | | 登録日 | 2023-07-12 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K2A
 
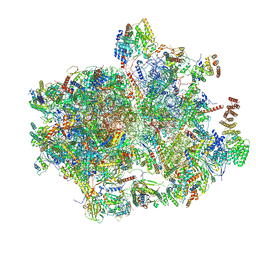 | | Cryo-EM structure of the human 55S mitoribosome with Tigecycline | | 分子名称: | 12S rRNA, 16S rRNA, 39S ribosomal protein L22, ... | | 著者 | Li, X, Wang, M, Cheng, J. | | 登録日 | 2023-07-12 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT2
 
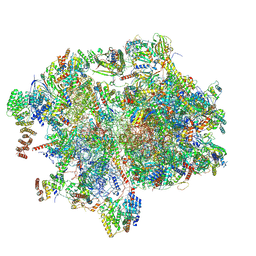 | | Cryo-EM structure of the human 55S mitoribosome with 10uM Tigecycline | | 分子名称: | 12s rRNA, 16s rRNA, 39S ribosomal protein L22, ... | | 著者 | Li, X, Wang, M, Cheng, J. | | 登録日 | 2024-01-10 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-11-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT1
 
 | | Cryo-EM structure of the human 39S mitoribosome with 5uM Tigecycline | | 分子名称: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | 著者 | Li, X, Wang, M, Cheng, J. | | 登録日 | 2024-01-10 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT0
 
 | | Cryo-EM structure of the human 55S mitoribosome with 5um Tigecycline | | 分子名称: | 12s rRNA, 16s rRNA, 39S ribosomal protein L22, ... | | 著者 | Li, X, Wang, M, Cheng, J. | | 登録日 | 2024-01-10 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XT3
 
 | | Cryo-EM structure of the human 39S mitoribosome with 10uM Tigecycline | | 分子名称: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | 著者 | Li, X, Wang, M, Cheng, J. | | 登録日 | 2024-01-10 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
6M40
 
 | | Crystal structure of the NS3-like helicase from Alongshan virus | | 分子名称: | NS3-like protein | | 著者 | Gao, X.P, Zhu, K.X, Chen, P, Wojdyla, J.A, Wang, M, Cui, S. | | 登録日 | 2020-03-05 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Crystal structure of the NS3-like helicase from Alongshan virus.
Iucrj, 7, 2020
|
|
6JB3
 
 | | Structure of SUR1 subunit bound with repaglinide | | 分子名称: | ATP-binding cassette sub-family C member 8 isoform X2, Digitonin, Repaglinide | | 著者 | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | 登録日 | 2019-01-25 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
7E6V
 
 | | The crystal structure of foot-and-mouth disease virus(FMDV) 2C protein 97-318aa | | 分子名称: | ACETATE ION, Protein 2C | | 著者 | Zhang, C, Wojdyla, J.A, Qin, B, Wang, M, Gao, X, Cui, S. | | 登録日 | 2021-02-24 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.832 Å) | | 主引用文献 | An anti-picornaviral strategy based on the crystal structure of foot-and-mouth disease virus 2C protein.
Cell Rep, 40, 2022
|
|
7CQ2
 
 | | Crystal structure of Slx1-Slx4 | | 分子名称: | GLYCEROL, SLX4 isoform 1, Structure-specific endonuclease subunit SLX1, ... | | 著者 | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | 登録日 | 2020-08-08 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
7CQ3
 
 | | Crystal structure of Slx1-Slx4 | | 分子名称: | SLX4 isoform 1, SULFATE ION, Structure-specific endonuclease subunit SLX1, ... | | 著者 | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | 登録日 | 2020-08-08 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.449 Å) | | 主引用文献 | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
7CQ4
 
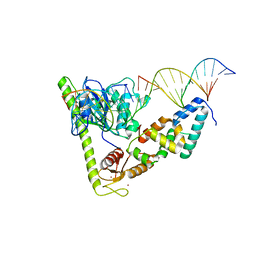 | | Crystal structure of Slx1-Slx4 in complex with 5'flap DNA | | 分子名称: | DNA (27-MER), DNA (5'-D(*AP*GP*GP*AP*CP*AP*TP*CP*TP*TP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*TP*AP*CP*AP*AP*CP*AP*GP*AP*T)-3'), ... | | 著者 | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | 登録日 | 2020-08-08 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.294 Å) | | 主引用文献 | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
