6MBL
 
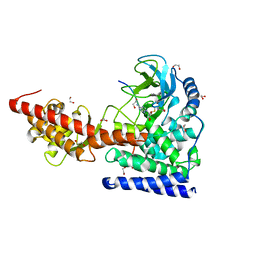 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, Second P212121 Crystal Form | | 分子名称: | 1,2-ETHANEDIOL, Actin Peptide, Histone-lysine N-methyltransferase setd3, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2018-08-30 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.197 Å) | | 主引用文献 | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MBJ
 
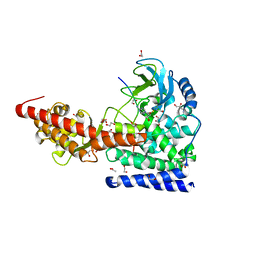 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, P21 Crystal Form | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | 著者 | Horton, J.R, Dai, S, Cheng, X. | | 登録日 | 2018-08-30 | | 公開日 | 2018-12-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
3OMH
 
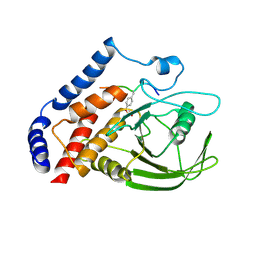 | | Crystal structure of PTPN22 in complex with SKAP-HOM pTyr75 peptide | | 分子名称: | Src kinase-associated phosphoprotein 2, Tyrosine-protein phosphatase non-receptor type 22 | | 著者 | Yu, X, Sun, J.-P, Zhang, S, Zhang, Z.-Y. | | 登録日 | 2010-08-26 | | 公開日 | 2011-06-29 | | 最終更新日 | 2011-09-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Substrate Specificity of Lymphoid-specific Tyrosine Phosphatase (Lyp) and Identification of Src Kinase-associated Protein of 55 kDa Homolog (SKAP-HOM) as a Lyp Substrate.
J.Biol.Chem., 286, 2011
|
|
7YGG
 
 | |
6KUR
 
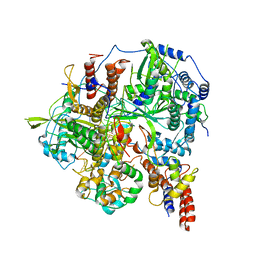 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUK
 
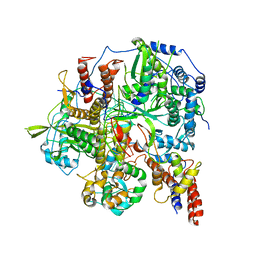 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUP
 
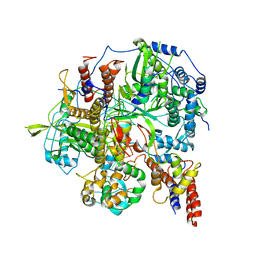 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUV
 
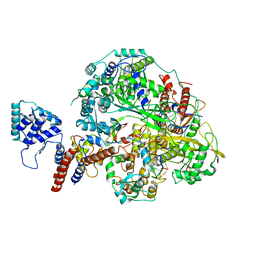 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | | 分子名称: | 3'-cRNA, 5'-cRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6UY0
 
 | |
6KUT
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | 分子名称: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-02 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KV5
 
 | | Structure of influenza D virus apo polymerase | | 分子名称: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | 著者 | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | 登録日 | 2019-09-03 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
8W2O
 
 | | Yeast U1 snRNP with humanized U1C Zinc-Finger domain | | 分子名称: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA, Pre-mRNA-processing factor 39, ... | | 著者 | Shi, S.S, Kuang, Z.L, Zhao, R. | | 登録日 | 2024-02-20 | | 公開日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Selected humanization of yeast U1 snRNP leads to global suppression of pre-mRNA splicing and mitochondrial dysfunction in the budding yeast.
Rna, 2024
|
|
8T9H
 
 | |
4Z16
 
 | | Crystal Structure of the Jak3 Kinase Domain Covalently Bound to N-(3-(((5-chloro-2-((2-methoxy-4-(4-methylpiperazin-1-yl)phenyl)amino)pyrimidin-4-yl)amino)methyl)phenyl)acrylamide | | 分子名称: | N-(3-{[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]methyl}phenyl)prop-2-enamide, Tyrosine-protein kinase JAK3 | | 著者 | McNally, R, Tan, L, Gray, N.S, Eck, M.J. | | 登録日 | 2015-03-26 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Development of Selective Covalent Janus Kinase 3 Inhibitors.
J.Med.Chem., 58, 2015
|
|
3VDP
 
 | |
3VE5
 
 | |
3VDU
 
 | |
8T9F
 
 | |
8THU
 
 | |
4PH4
 
 | | The crystal structure of Human VPS34 in complex with PIK-III | | 分子名称: | 4'-(cyclopropylmethyl)-N~2~-(pyridin-4-yl)-4,5'-bipyrimidine-2,2'-diamine, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | 著者 | Knapp, M.S, Elling, R.A. | | 登録日 | 2014-05-04 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo.
Nat.Cell Biol., 16, 2014
|
|
8IFG
 
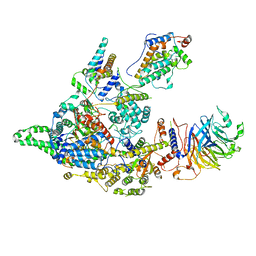 | | Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe | | 分子名称: | Chromatin modification-related protein eaf3, Cph1, Cph2, ... | | 著者 | Zhang, H.Q, Wang, X, Wang, Y.N, Liu, S.M, Zhang, Y, Xu, K, Ji, L.T, Kornberg, R.D. | | 登録日 | 2023-02-17 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Class I histone deacetylase complex: Structure and functional correlates.
Proc Natl Acad Sci U S A, 120, 2023
|
|
5XTA
 
 | | Crystal structure of lpg1832, a VirK family protein from Legionella pneumophila | | 分子名称: | GLYCEROL, SULFATE ION, VirK protein | | 著者 | Yin, S, Gong, X, Zhang, N, Ge, H. | | 登録日 | 2017-06-18 | | 公開日 | 2017-08-16 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of lpg1832, a VirK family protein from Legionella pneumophila, reveals a novel fold for bacterial VirK proteins
FEBS Lett., 591, 2017
|
|
7CMZ
 
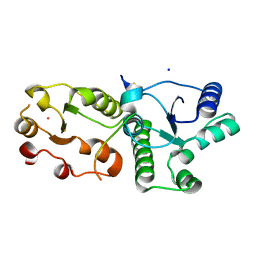 | | Crystal Structure of BRCT7/8 in Complex with the APS Motif of PHF8 | | 分子名称: | DNA topoisomerase 2-binding protein 1, Histone lysine demethylase PHF8, POTASSIUM ION, ... | | 著者 | Che, S.Y, Ma, S, Cao, C, Yao, Z, Shi, L, Yang, N. | | 登録日 | 2020-07-29 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.695 Å) | | 主引用文献 | PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability.
Sci Adv, 7, 2021
|
|
3EIX
 
 | |
3EIW
 
 | |
