7F8X
 
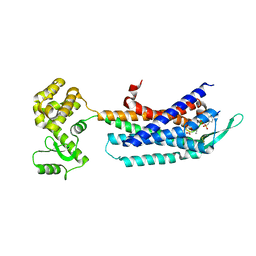 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with NN9056 | | 分子名称: | ASP-SMF-NLE-GLY-TRP-NLE-OEM-MEA-NH2 (NN9056), Cholecystokinin receptor type A,Endolysin | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
6A4M
 
 | |
7F8Y
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with devazepide | | 分子名称: | N-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-1H-indole-2-carboxamide, fusion protein of Cholecystokinin receptor type A and Endolysin | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | 分子名称: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8W
 
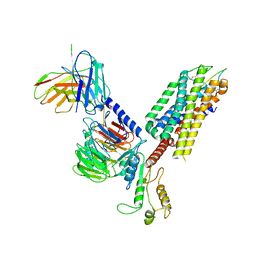 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | 分子名称: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-13 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8V
 
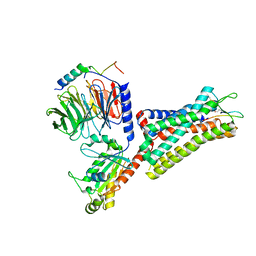 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | 分子名称: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | 登録日 | 2021-07-02 | | 公開日 | 2021-10-13 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F3P
 
 | | Crystal structure of a nadp-dependent alcohol dehydrogenase mutant in apo form | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent isopropanol dehydrogenase, ZINC ION | | 著者 | Han, X, Bi, Y, Wei, H.L, Gao, J, Li, Q, Qu, G, Sun, Z.T, Liu, W.D. | | 登録日 | 2021-06-16 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Unlocking the Stereoselectivity and Substrate Acceptance of Enzymes: Proline-Induced Loop Engineering Test.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7EPT
 
 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | 分子名称: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | 登録日 | 2021-04-27 | | 公開日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
8YQP
 
 | |
8YQJ
 
 | | Crystal structure of HylD1 | | 分子名称: | Lipase | | 著者 | Wang, N, Li, C.Y. | | 登録日 | 2024-03-19 | | 公開日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Molecular insights into the catalytic mechanism of a phthalate ester hydrolase.
J Hazard Mater, 476, 2024
|
|
4HVB
 
 | | Catalytic unit of PI3Kg in complex with PI3K/mTOR dual inhibitor PF-04979064 | | 分子名称: | 1-{1-[(2S)-2-hydroxypropanoyl]piperidin-4-yl}-3-methyl-8-(6-methylpyridin-3-yl)-1,3-dihydro-2H-imidazo[4,5-c][1,5]naphthyridin-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | 著者 | Knighton, D.R, Cheng, H. | | 登録日 | 2012-11-05 | | 公開日 | 2013-11-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Discovery of the Highly Potent PI3K/mTOR Dual Inhibitor PF-04979064 through Structure-Based Drug Design.
ACS Med Chem Lett, 4, 2013
|
|
4JOL
 
 | |
8X17
 
 | | Cryo-EM structure of adenosine receptor A3AR bound to CF102 | | 分子名称: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Cai, H, Xu, Y, Xu, H.E. | | 登録日 | 2023-11-06 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Cryo-EM structures of adenosine receptor A 3 AR bound to selective agonists.
Nat Commun, 15, 2024
|
|
8X16
 
 | | Cryo-EM structure of adenosine receptor A3AR bound to CF101 | | 分子名称: | Adenosine receptor A3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Cai, H, Xu, Y, Xu, H.E. | | 登録日 | 2023-11-06 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Cryo-EM structures of adenosine receptor A 3 AR bound to selective agonists.
Nat Commun, 15, 2024
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | 分子名称: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | 著者 | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | 登録日 | 2020-09-23 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7X3Y
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (CVB1-E:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-01 | | 公開日 | 2023-06-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X38
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 8A10 (CVB1-E:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3F
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 9A3 (CVB1-A:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X42
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP0, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-01 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2G
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb nAb 2E6 (CVB1-E:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2I
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 2E6 (CVB1-pre-A:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2O
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 2E6 (CVB1-M:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-25 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X2W
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A particle in complex with nAb 8A10 (CVB1-pre-A:8A10) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2022-10-05 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
