6YES
 
 | |
6LR6
 
 | | The crystal structure of human cytoplasmic LRS | | 分子名称: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | 著者 | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | 登録日 | 2020-01-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.009 Å) | | 主引用文献 | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6S8B
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1 | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.41 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S91
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2 | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-11 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SIC
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr1 family, Cmr4 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S8E
 
 | | Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SH8
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2, in the presence of ssDNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SHB
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1, in the presence of ssDNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6OG2
 
 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, post-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-04-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OAX
 
 | | Structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Hyperactive disaggregase ClpB, ... | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-03-18 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OG3
 
 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, NTD-trimer | | 分子名称: | Alpha S1-casein, Hyperactive disaggregase ClpB | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-04-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OAY
 
 | | Structure of the hyperactive ClpB mutant K476C, bound to casein, post-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Hyperactive disaggregase ClpB, ... | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-03-18 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6S6B
 
 | | Type III-B Cmr-beta Cryo-EM structure of the Apo state | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-02 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
4U93
 
 | | Crystal Structure of Hsp90-alpha N-domain Bound to the Inhibitor NVP-HSP990 | | 分子名称: | (7R)-2-amino-7-[4-fluoro-2-(6-methoxypyridin-2-yl)phenyl]-4-methyl-7,8-dihydropyrido[4,3-d]pyrimidin-5(6H)-one, Heat shock protein HSP 90-alpha | | 著者 | Bellamacina, C.R, Shafer, C.M, Bussiere, D. | | 登録日 | 2014-08-05 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Design, Structure-Activity Relationship, and in Vivo Characterization of the Development Candidate NVP-HSP990.
J.Med.Chem., 57, 2014
|
|
1VHN
 
 | |
6O9H
 
 | | Mouse ECD with Fab1 | | 分子名称: | Gastric inhibitory polypeptide receptor, Heavy chain, Light chain, ... | | 著者 | Min, X, Wang, Z. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-22 | | 最終更新日 | 2020-04-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular mechanism of an antagonistic antibody against glucose-dependent insulinotropic polypeptide receptor.
Mabs, 12, 2020
|
|
6RVA
 
 | | STRUCTURE OF [ASP58]-IGF-I ANALOGUE | | 分子名称: | Insulin-like growth factor I | | 著者 | Jiracek, J, Zakova, L, Socha, O. | | 登録日 | 2019-05-31 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mutations at hypothetical binding site 2 in insulin and insulin-like growth factors 1 and 2 result in receptor- and hormone-specific responses.
J.Biol.Chem., 294, 2019
|
|
4WQ6
 
 | | The crystal structure of human Nicotinamide phosphoribosyltransferase (NAMPT) in complex with N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide inhibitor (compound 21) | | 分子名称: | 1,2-ETHANEDIOL, N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Li, D, Wang, W. | | 登録日 | 2014-10-21 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Identification of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with no evidence of CYP3A4 time-dependent inhibition and improved aqueous solubility.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1VI7
 
 | |
6XTI
 
 | | NMR solution structure of class IV lasso peptide felipeptin A2 from Amycolatopsis sp. YIM10 | | 分子名称: | Felipeptin A2 | | 著者 | Madland, E, Aachmann, F.L, Guerrero-Garzon, J.F, Zotchev, S.B, Courtade, G. | | 登録日 | 2020-01-16 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
6XTH
 
 | | NMR solution structure of class IV lasso peptide felipeptin A1 from Amycolatopsis sp. YIM10 | | 分子名称: | Felipeptin A1 | | 著者 | Madland, E, Guerrero-Garzon, J.F, Zotchev, S.B, Aachmann, F.L, Courtade, G. | | 登録日 | 2020-01-16 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
6N33
 
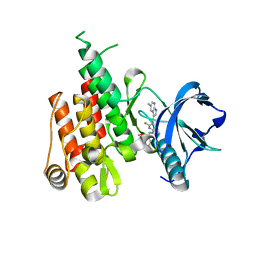 | | Crystal structure of fms kinase domain with a small molecular inhibitor, PLX5622 | | 分子名称: | 6-fluoro-N-[(5-fluoro-2-methoxypyridin-3-yl)methyl]-5-[(5-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | 著者 | Zhang, Y. | | 登録日 | 2018-11-14 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer's disease model.
Nat Commun, 10, 2019
|
|
8HO4
 
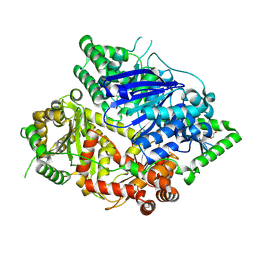 | | Falcilysin in complex with MMV000848 | | 分子名称: | (2R)-1-(9H-carbazol-9-yl)-3-(cyclopentylamino)propan-2-ol, 1,2-ETHANEDIOL, Falcilysin, ... | | 著者 | Lin, J.Q, Chung, Z, Lescar, J. | | 登録日 | 2022-12-09 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Identification of an inhibitory pocket in falcilysin provides a new avenue for malaria drug development.
Cell Chem Biol, 31, 2024
|
|
8HO5
 
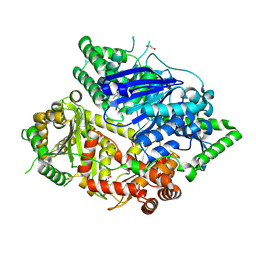 | | Falcilysin in complex with MMV665806 | | 分子名称: | (2S)-1-(2,3-dimethyl-1H-indol-1-yl)-3-{[(1S,2S)-2-methylcyclohexyl]amino}propan-2-ol, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Lin, J.Q, Chung, Z, Lescar, J. | | 登録日 | 2022-12-09 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Identification of an inhibitory pocket in falcilysin provides a new avenue for malaria drug development.
Cell Chem Biol, 31, 2024
|
|
4S0Z
 
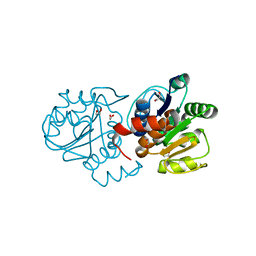 | | Crystal structure of M26V human DJ-1 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Protein DJ-1 | | 著者 | Milkovic, N.M, Wilson, M.A. | | 登録日 | 2015-01-07 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Transient sampling of aggregation-prone conformations causes pathogenic instability of a parkinsonian mutant of DJ-1 at physiological temperature.
Protein Sci., 24, 2015
|
|
