8OYX
 
 | |
8OYW
 
 | |
8OYY
 
 | |
6P6F
 
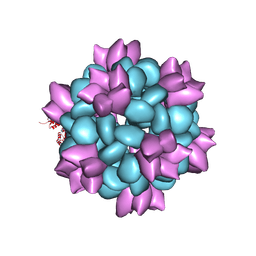 | | BG505 SOSIP-I53-50NP | | 分子名称: | I53-50A.1NT1, I53-50B.4PT1 | | 著者 | Berndsen, Z.T, Ward, A.B. | | 登録日 | 2019-06-03 | | 公開日 | 2019-06-19 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle.
Nat Commun, 10, 2019
|
|
5JSB
 
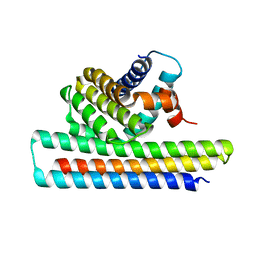 | | Crystal structure of Mcl1-inhibitor complex | | 分子名称: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 inhibitor | | 著者 | Shen, B.W, Stoddard, B.L. | | 登録日 | 2016-05-07 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer.
Elife, 5, 2016
|
|
3B5L
 
 | |
2A0M
 
 | |
4RZT
 
 | | Lac repressor engineered to bind sucralose, sucralose-bound tetramer | | 分子名称: | 4-chloro-4-deoxy-alpha-D-galactopyranose-(1-2)-1,6-dichloro-1,6-dideoxy-beta-D-fructofuranose, Lac repressor | | 著者 | Arbing, M.A, Cascio, D, Sawaya, M.R, Kosuri, S, Church, G.M. | | 登録日 | 2014-12-24 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
4RZS
 
 | | Lac repressor engineered to bind sucralose, unliganded tetramer | | 分子名称: | GLYCEROL, Lac repressor | | 著者 | Arbing, M.A, Cascio, D, Kosuri, S, Church, G.M. | | 登録日 | 2014-12-24 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Engineering an allosteric transcription factor to respond to new ligands.
Nat.Methods, 13, 2016
|
|
6OHH
 
 | | Structure of EF1p2_mFAP2b bound to DFHBI | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, CALCIUM ION, EF1p2_mFAP2b | | 著者 | Doyle, L.A, Stoddard, B.L. | | 登録日 | 2019-04-05 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Nat Commun, 12, 2021
|
|
5IEN
 
 | | Structure of CDL2.2, a computationally designed Vitamin-D3 binder | | 分子名称: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.2, GLYCEROL | | 著者 | Stoddard, B.L, Doyle, L.A. | | 登録日 | 2016-02-25 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.089 Å) | | 主引用文献 | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
6S1V
 
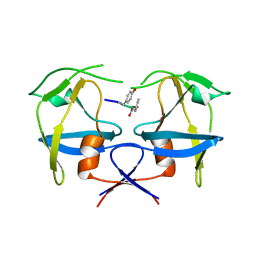 | | Crystal structure of dimeric M-PMV protease D26N mutant in complex with inhibitor | | 分子名称: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5AN7
 
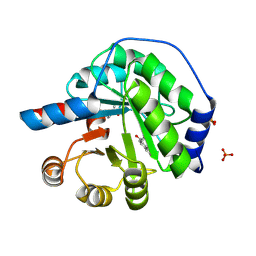 | | Structure of the engineered retro-aldolase RA95.5-8F with a bound 1,3-diketone inhibitor | | 分子名称: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, PHOSPHATE ION, RA95.5-8F | | 著者 | Obexer, R, Mittl, P.R.E, Hilvert, D. | | 登録日 | 2015-09-04 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
5IEO
 
 | | Structure of CDL2.3a, a computationally designed Vitamin-D3 binder | | 分子名称: | 1,2-ETHANEDIOL, 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, CDL2.3a | | 著者 | Stoddard, B.L, Doyle, L.A. | | 登録日 | 2016-02-25 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Unintended specificity of an engineered ligand-binding protein facilitated by unpredicted plasticity of the protein fold.
Protein Eng.Des.Sel., 31, 2018
|
|
5AOU
 
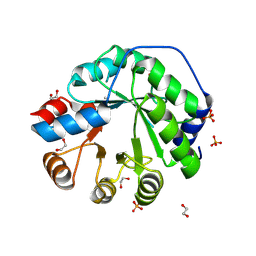 | | Structure of the engineered retro-aldolase RA95.5-8F apo | | 分子名称: | 1,2-ETHANEDIOL, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, PHOSPHATE ION | | 著者 | Obexer, R, Mittl, P, Hilvert, D. | | 登録日 | 2015-09-11 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
5IEP
 
 | |
5IF6
 
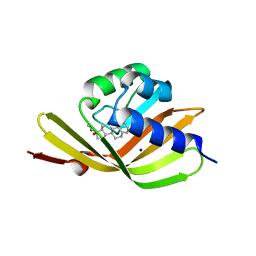 | |
7UNJ
 
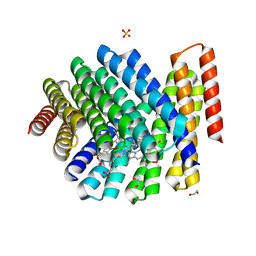 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | 分子名称: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | 著者 | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | 登録日 | 2022-04-11 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
7UNH
 
 | |
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | 著者 | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | 登録日 | 2022-04-11 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
6S1U
 
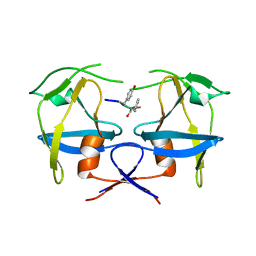 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | 分子名称: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1W
 
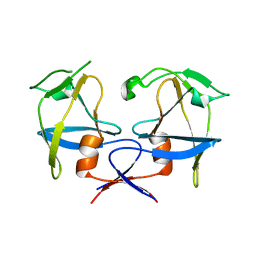 | | Crystal structure of dimeric M-PMV protease D26N mutant | | 分子名称: | Gag-Pro-Pol polyprotein | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5IER
 
 | |
5JG9
 
 | |
3U0S
 
 | | Crystal Structure of an Enzyme Redesigned Through Multiplayer Online Gaming: CE6 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Diisopropyl-fluorophosphatase, GLYCEROL, ... | | 著者 | Bale, J.B, Shen, B.W, Stoddard, B.L. | | 登録日 | 2011-09-29 | | 公開日 | 2012-02-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Increased Diels-Alderase activity through backbone remodeling guided by Foldit players.
Nat.Biotechnol., 30, 2012
|
|
