6P6B
 
 | |
6VTI
 
 | | Solution NMR structure of the N-terminal domain of the Serine/threonine-protein phosphatase 1 regulatory subunit 10, PPP1R10 | | 分子名称: | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | | 著者 | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | 登録日 | 2020-02-12 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
6BY9
 
 | | Crystal structure of EHMT1 | | 分子名称: | Histone-lysine N-methyltransferase EHMT1, UNKNOWN ATOM OR ION | | 著者 | Dong, A, Wei, Y, Li, A, Tempel, W, Han, S, Sunnerhagen, M, Penn, L, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2017-12-20 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of EHMT1
to be published
|
|
4XO2
 
 | |
1S14
 
 | |
1S16
 
 | |
4EFB
 
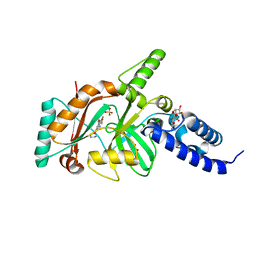 | | Crystal structure of DNA ligase | | 分子名称: | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | 著者 | Wei, Y, Wang, T, Charifson, P, Xu, W. | | 登録日 | 2012-03-29 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of DNA ligase
To be Published
|
|
4EFE
 
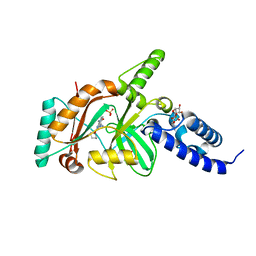 | | crystal structure of DNA ligase | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | 著者 | Wei, Y, Wang, T, Charifson, P, Xu, W. | | 登録日 | 2012-03-29 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | crystal structure of DNA ligase
To be Published
|
|
4F1R
 
 | |
7LQT
 
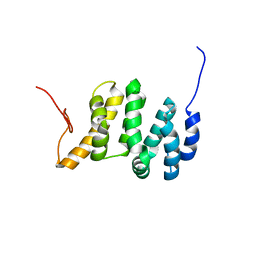 | | Solution NMR structure of the PNUTS amino-terminal Domain fused to Myc Homology Box 0 | | 分子名称: | Serine/threonine-protein phosphatase 1 regulatory subunit 10,Myc proto-oncogene protein fusion | | 著者 | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | 登録日 | 2021-02-15 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
3TKA
 
 | | crystal structure and solution saxs of methyltransferase rsmh from E.coli | | 分子名称: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Ribosomal RNA small subunit methyltransferase H, S-ADENOSYLMETHIONINE, ... | | 著者 | Gao, Z.Q, Wei, Y, Zhang, H, Dong, Y.H. | | 登録日 | 2011-08-25 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal and solution structures of methyltransferase RsmH provide basis for methylation of C1402 in 16S rRNA.
J.Struct.Biol., 179, 2012
|
|
3B92
 
 | |
3G7B
 
 | |
3G75
 
 | |
3G7E
 
 | |
2OIN
 
 | | crystal structure of HCV NS3-4A R155K mutant | | 分子名称: | NS4A peptide, Polyprotein, ZINC ION | | 著者 | Wei, Y. | | 登録日 | 2007-01-11 | | 公開日 | 2007-06-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha.
J.Biol.Chem., 282, 2007
|
|
7E7Y
 
 | |
2OI0
 
 | | Crystal structure analysis 0f the TNF-a Coverting Enzyme (TACE) in complexed with Aryl-sulfonamide | | 分子名称: | (3S)-1-{[4-(BUT-2-YN-1-YLOXY)PHENYL]SULFONYL}PYRROLIDINE-3-THIOL, TNF- a Converting Enzyme (TACE), ZINC ION | | 著者 | Wei, Y, Rao, G.B, Bandarage, U.K. | | 登録日 | 2007-01-10 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Novel thiol-based TACE inhibitors: rational design, synthesis, and SAR of thiol-containing aryl sulfonamides
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2P59
 
 | | Crystal Structure of Hepatitis C Virus NS3.4A protease | | 分子名称: | (2S,3AS,7AS)-1-[(2S)-2-{[(2S)-2-CYCLOHEXYL-2-({[(2R)-4-NITRO-2H-PYRROL-2-YL]CARBONYL}AMINO)ACETYL]AMINO}-3,3-DIMETHYLBUTANOYL]-N-{(1S)-1-[(1R)-2-(CYCLOPROPYLAMINO)-1-HYDROXY-2-OXOETHYL]BUTYL}OCTAHYDRO-1H-INDOLE-2-CARBOXAMIDE, NS3, peptide | | 著者 | Perni, R.B, Wei, Y. | | 登録日 | 2007-03-14 | | 公開日 | 2008-02-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Inhibitors of hepatitis C virus NS3.4A protease. Effect of P4 capping groups on inhibitory potency and pharmacokinetics.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5MV1
 
 | | Crystal structure of the E protein of the Japanese encephalitis virulent virus | | 分子名称: | E protein | | 著者 | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | 登録日 | 2017-01-14 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
5MV2
 
 | | Crystal structure of the E protein of the Japanese encephalitis live attenuated vaccine virus | | 分子名称: | E protein | | 著者 | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | 登録日 | 2017-01-14 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
1TIB
 
 | |
6L62
 
 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | 分子名称: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | 著者 | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | 登録日 | 2019-10-25 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-04-29 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
1WAB
 
 | | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | 分子名称: | ACETATE ION, PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | 著者 | Ho, Y.S, Swenson, L, Derewenda, U, Serre, L, Wei, Y, Dauter, Z, Hattori, M, Aoki, J, Arai, H, Adachi, T, Inoue, K, Derewenda, Z.S. | | 登録日 | 1996-10-30 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer.
Nature, 385, 1997
|
|
1TIA
 
 | | AN UNUSUAL BURIED POLAR CLUSTER IN A FAMILY OF FUNGAL LIPASES | | 分子名称: | LIPASE | | 著者 | Derewenda, U, Swenson, L, Yamaguchi, S, Wei, Y, Derewenda, Z.S. | | 登録日 | 1993-12-06 | | 公開日 | 1995-01-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | An unusual buried polar cluster in a family of fungal lipases.
Nat.Struct.Biol., 1, 1994
|
|
