1JG9
 
 | |
1JGI
 
 | |
8B2G
 
 | | SH3-like domain from Penicillium virgatum muramidase | | 分子名称: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | 著者 | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | 登録日 | 2022-09-13 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2F
 
 | | SH3-like cell wall binding domain of the GH24 family muramidase from Trichophaea saccata in complex with triglycine | | 分子名称: | 1,2-ETHANEDIOL, GLY-GLY-GLY, SH3-like cell wall binding domain-containing protein, ... | | 著者 | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | 登録日 | 2022-09-13 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.183 Å) | | 主引用文献 | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2E
 
 | | Muramidase from Kionochaeta sp natural catalytic core | | 分子名称: | CADMIUM ION, Muramidase | | 著者 | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | 登録日 | 2022-09-13 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2H
 
 | | Muramidase from Thermothielavioides terrestris, catalytic domain | | 分子名称: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | 著者 | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | 登録日 | 2022-09-13 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2S
 
 | | GH24 family muramidase from Trichophaea saccata with an SH3-like cell wall binding domain | | 分子名称: | GH24 family muramidase, POTASSIUM ION | | 著者 | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | 登録日 | 2022-09-14 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1XWV
 
 | | Structure of the house dust mite allergen Der f 2: Implications for function and molecular basis of IgE cross-reactivity | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Der f II | | 著者 | Johannessen, B.R, Skov, L.K, Kastrup, J.S, Kristensen, O, Bolwig, C, Larsen, J.N, Spangfort, M, Lund, K, Gajhede, M. | | 登録日 | 2004-11-02 | | 公開日 | 2004-12-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structure of the house dust mite allergen Der f 2: implications for function and molecular basis of IgE cross-reactivity.
Febs Lett., 579, 2005
|
|
1YCJ
 
 | | Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate | | 分子名称: | GLUTAMIC ACID, Ionotropic glutamate receptor 5, SULFATE ION | | 著者 | Naur, P, Vestergaard, B, Skov, L.K, Egebjerg, J, Gajhede, M, Kastrup, J.S. | | 登録日 | 2004-12-22 | | 公開日 | 2005-02-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate
Febs Lett., 579, 2005
|
|
6FHV
 
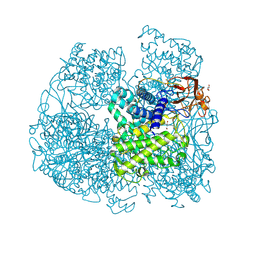 | | Crystal structure of Penicillium oxalicum Glucoamylase | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | 登録日 | 2018-01-15 | | 公開日 | 2018-05-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FRV
 
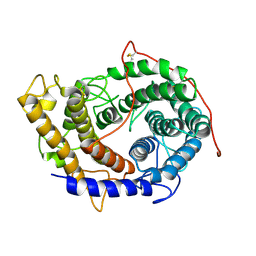 | | Structure of the catalytic domain of Aspergillus niger Glucoamylase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucoamylase, ... | | 著者 | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | 登録日 | 2018-02-16 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FHW
 
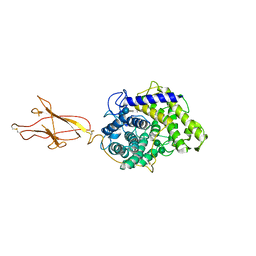 | | Structure of Hormoconis resinae Glucoamylase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Roth, C, Moroz, O.V, Ariza, A, Friis, E.P, Davies, G.J, Wilson, K.S. | | 登録日 | 2018-01-15 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural insight into industrially relevant glucoamylases: flexible positions of starch-binding domains.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4YMB
 
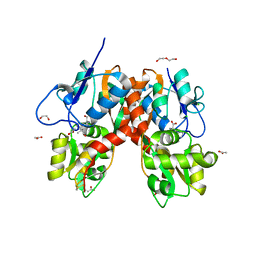 | | Structure of the ligand-binding domain of GluK1 in complex with the antagonist CNG10111 | | 分子名称: | (3R,4S)-3-(3-carboxyphenyl)-4-propyl-L-proline, 1,2-ETHANEDIOL, ACETATE ION, ... | | 著者 | Moller, C, Tapken, D, Kastrup, J.S, Frydenvang, K. | | 登録日 | 2015-03-06 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure-Activity Relationship Study of Ionotropic Glutamate Receptor Antagonist (2S,3R)-3-(3-Carboxyphenyl)pyrrolidine-2-carboxylic Acid.
J.Med.Chem., 58, 2015
|
|
2PBW
 
 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex with the Partial agonist Domoic Acid at 2.5 A Resolution | | 分子名称: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, Glutamate receptor, ionotropic kainate 1 | | 著者 | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-03-29 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
5MFW
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-121 at 2.10 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | 著者 | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
5MFQ
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-344 at 1.90 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | 著者 | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
5M2V
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (2S,4R)-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid at 3.18 A resolution | | 分子名称: | (2~{S},4~{R})-4-(2-carboxyphenoxy)pyrrolidine-2-carboxylic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Frydenvang, K, Kastrup, J.S, Kristensen, C.M. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Design and Synthesis of a Series of l-trans-4-Substituted Prolines as Selective Antagonists for the Ionotropic Glutamate Receptors Including Functional and X-ray Crystallographic Studies of New Subtype Selective Kainic Acid Receptor Subtype 1 (GluK1) Antagonist (2S,4R)-4-(2-Carboxyphenoxy)pyrrolidine-2-carboxylic Acid.
J. Med. Chem., 60, 2017
|
|
5MFV
 
 | | Crystal structure of the GluK1 ligand-binding domain in complex with kainate and BPAM-521 at 2.18 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-Cyclopropyl-3,4-dihydro-7-hydroxy-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, ... | | 著者 | Larsen, A.P, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2016-11-18 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.181 Å) | | 主引用文献 | Identification and Structure-Function Study of Positive Allosteric Modulators of Kainate Receptors.
Mol. Pharmacol., 91, 2017
|
|
1VSO
 
 | | Crystal Structure of the Ligand-Binding Core of iGluR5 in Complex With the Antagonist (S)-ATPO at 1.85 A resolution | | 分子名称: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, GLYCEROL, Glutamate receptor, ... | | 著者 | Hald, H, Naur, P, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-03-29 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Partial agonism and antagonism of the ionotropic glutamate receptor iGLuR5: structures of the ligand-binding core in complex with domoic acid and 2-amino-3-[5-tert-butyl-3-(phosphonomethoxy)-4-isoxazolyl]propionic acid.
J.Biol.Chem., 282, 2007
|
|
4DLD
 
 | | Crystal structure of the GluK1 ligand-binding domain (S1S2) in complex with the antagonist (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid at 2.0 A resolution | | 分子名称: | (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2012-02-06 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and pharmacological characterization of phenylalanine-based AMPA receptor antagonists at kainate receptors
Chemmedchem, 7, 2012
|
|
4E0X
 
 | |
3S2V
 
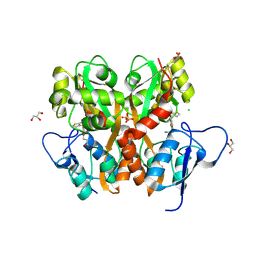 | | Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution | | 分子名称: | (S)-1-(2'-AMINO-2'-CARBOXYETHYL)-3-[(2-CARBOXYTHIEN-3-YL)METHYL]THIENO[3,4-D]PYRIMIDIN-2,4-DIONE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2011-05-17 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization.
J.Med.Chem., 54, 2011
|
|
3GBA
 
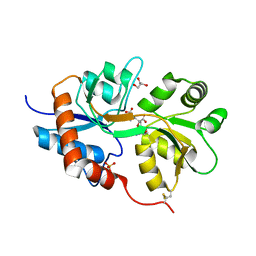 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with dysiherbaine at 1.35A resolution | | 分子名称: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-02-19 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
3GBB
 
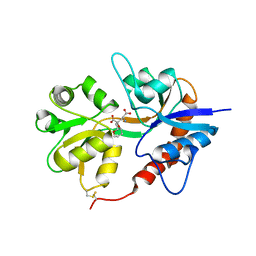 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with MSVIII-19 at 2.10A resolution | | 分子名称: | (2R,3aR,7aR)-2-[(2S)-2-amino-3-hydroxy-3-oxo-propyl]-3,3a,5,6,7,7a-hexahydrofuro[4,5-b]pyran-2-carboxylic acid, Glutamate receptor, ionotropic kainate 1 | | 著者 | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | 登録日 | 2009-02-19 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
