1PWM
 
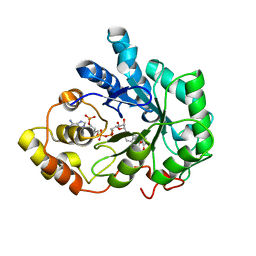 | | Crystal structure of human Aldose Reductase complexed with NADP and Fidarestat | | 分子名称: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | El-Kabbani, O, Darmanin, C, Schneider, T.R, Hazemann, I, Ruiz, F, Oka, M, Joachimiak, A, Schulze-Briese, C, Tomizaki, T, Mitschler, A, Podjarny, A. | | 登録日 | 2003-07-02 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (0.92 Å) | | 主引用文献 | Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors
PROTEINS, 55, 2004
|
|
7LYZ
 
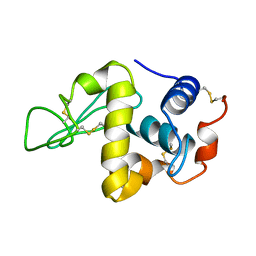 | | PROTEIN MODEL BUILDING BY THE USE OF A CONSTRAINED-RESTRAINED LEAST-SQUARES PROCEDURE | | 分子名称: | HEN EGG WHITE LYSOZYME | | 著者 | Moult, J, Yonath, A, Sussman, J, Herzberg, O, Podjarny, A, Traub, W. | | 登録日 | 1977-05-06 | | 公開日 | 1977-06-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Protein Model Building by the Use of a Constrained-Restrained Least-Squares Procedure
J.Appl.Crystallogr., 16, 1983
|
|
5EUC
 
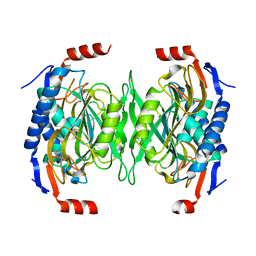 | | The role of the C-terminal region on the oligomeric state and enzymatic activity of Trypanosoma cruzi hypoxanthine phosphoribosyl transferase | | 分子名称: | Hypoxanthine-guanine phosphoribosyltransferase | | 著者 | Valsecchi, W.M, Cousido-Siah, A, Mitschler, A, Podjarny, A, Delfino, J.M, Santos, J. | | 登録日 | 2015-11-18 | | 公開日 | 2016-03-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | The role of the C-terminal region on the oligomeric state and enzymatic activity of Trypanosoma cruzi hypoxanthine phosphoribosyl transferase.
Biochim.Biophys.Acta, 1864, 2016
|
|
3PP6
 
 | | REP1-NXSQ fatty acid transporter Y128F mutant | | 分子名称: | PALMITOLEIC ACID, ReP1-NCXSQ | | 著者 | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | 登録日 | 2010-11-24 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3PPT
 
 | | REP1-NXSQ fatty acid transporter | | 分子名称: | PALMITOLEIC ACID, ReP1-NCXSQ | | 著者 | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | 登録日 | 2010-11-25 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.28 Å) | | 主引用文献 | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5NRA
 
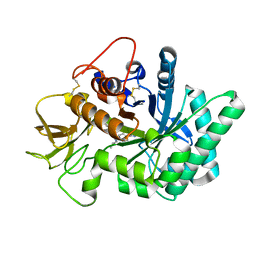 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7g | | 分子名称: | 1-(5-azanyl-4~{H}-1,2,4-triazol-3-yl)-~{N}-[2-(4-bromophenyl)ethyl]-~{N}-(2-methylpropyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | 著者 | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | 登録日 | 2017-04-22 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.267 Å) | | 主引用文献 | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
2I17
 
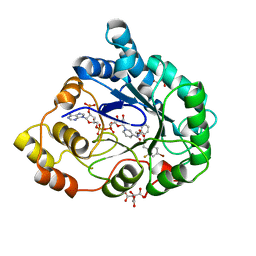 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594 at temperature of 60K | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Mitshler, A, Hasemann, I, Schneider, T, Cousido, A, Lunin, V.Y, Joachimiak, A, Podjarny, A. | | 登録日 | 2006-08-13 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.81 Å) | | 主引用文献 | Ultrahigh-resolution study of protein atomic displacement parameters at cryotemperatures obtained with a helium cryostat.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3QF6
 
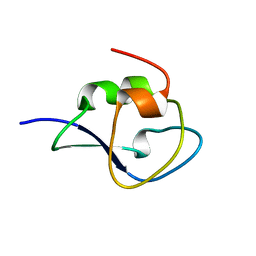 | | Neutron structure of type-III Antifreeze Protein allows the reconstruction of AFP-ice interface | | 分子名称: | Type-3 ice-structuring protein HPLC 12 | | 著者 | Howard, E.I, Blakeley, M.P, Haertlein, M, Petit-Haertlein, I, Mitschler, A, Fisher, S.J, Cousido-Siah, A, Salvay, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A. | | 登録日 | 2011-01-21 | | 公開日 | 2011-06-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å) | | 主引用文献 | Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface.
J.Mol.Recognit., 24, 2011
|
|
2I16
 
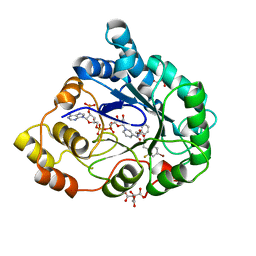 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594 at temperature of 15K | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Mitshler, A, Hasemann, I, Schneider, T, Cousido, A, Lunin, V.Y, Joachimiak, A, Podjarny, A. | | 登録日 | 2006-08-13 | | 公開日 | 2006-08-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.81 Å) | | 主引用文献 | Ultrahigh-resolution study of protein atomic displacement parameters at cryotemperatures obtained with a helium cryostat.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1AH4
 
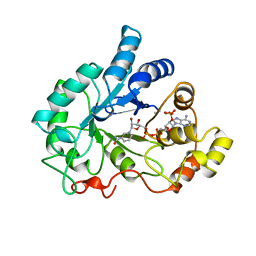 | | PIG ALDOSE REDUCTASE, HOLO FORM | | 分子名称: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Moras, D, Podjarny, A. | | 登録日 | 1997-04-12 | | 公開日 | 1998-04-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A 'specificity' pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil.
Structure, 5, 1997
|
|
5HR2
 
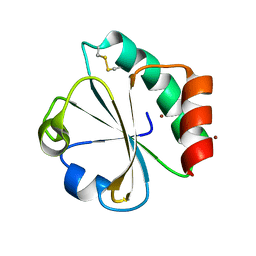 | | Crystal structure of thioredoxin L94A mutant | | 分子名称: | COPPER (II) ION, Thioredoxin | | 著者 | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | 登録日 | 2016-01-22 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
5HR0
 
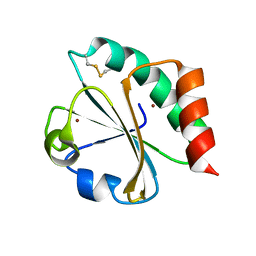 | | Crystal structure of thioredoxin E101G mutant | | 分子名称: | COPPER (II) ION, Thioredoxin | | 著者 | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | 登録日 | 2016-01-22 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
5HR3
 
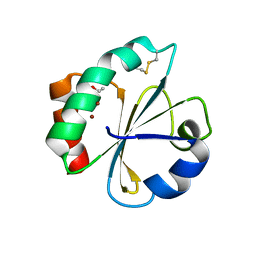 | | Crystal structure of thioredoxin N106A mutant | | 分子名称: | COPPER (II) ION, ETHANOL, SULFATE ION, ... | | 著者 | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | 登録日 | 2016-01-22 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.101 Å) | | 主引用文献 | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
5HBF
 
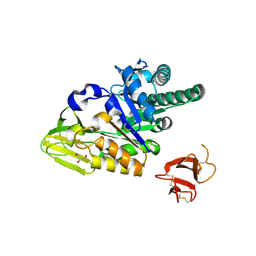 | | Crystal structure of human full-length chitotriosidase (CHIT1) | | 分子名称: | Chitotriosidase-1, GLYCEROL | | 著者 | Fadel, F, Zhao, Y, Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | 登録日 | 2015-12-31 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | X-Ray Crystal Structure of the Full Length Human Chitotriosidase (CHIT1) Reveals Features of Its Chitin Binding Domain.
Plos One, 11, 2016
|
|
5HR1
 
 | | Crystal structure of thioredoxin L107A mutant | | 分子名称: | COPPER (II) ION, Thioredoxin-1 | | 著者 | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | 登録日 | 2016-01-22 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.144 Å) | | 主引用文献 | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
6SJV
 
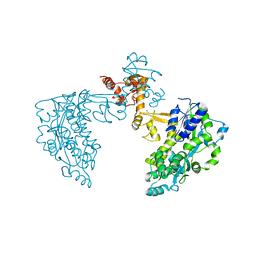 | | Structure of HPV18 E6 oncoprotein in complex with mutant E6AP LxxLL motif | | 分子名称: | Maltodextrin-binding protein,Protein E6,Ubiquitin-protein ligase E3A, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Kostmann, C, Mitschler, A, Podjarny, A, Trave, G. | | 登録日 | 2019-08-14 | | 公開日 | 2019-09-04 | | 最終更新日 | 2022-03-30 | | 実験手法 | X-RAY DIFFRACTION (2.029 Å) | | 主引用文献 | Cellular target recognition by HPV18 and HPV49 oncoproteins
To be published
|
|
6SJA
 
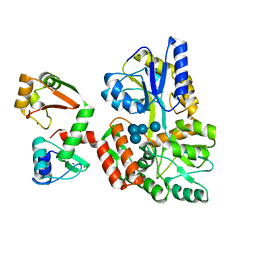 | | Structure of HPV16 E6 oncoprotein in complex with IRF3 LxxLL motif | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | 著者 | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | 登録日 | 2019-08-13 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Deciphering de molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
6SMV
 
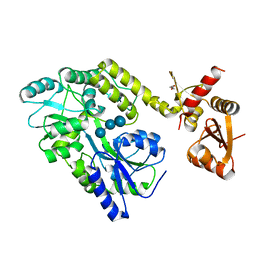 | | Structure of HPV49 E6 protein in complex with MAML1 LxxLL motif | | 分子名称: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Mastermind-like protein 1, ZINC ION, ... | | 著者 | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Kostmann, C, Mitschler, A, Podjarny, A, Trave, G. | | 登録日 | 2019-08-22 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Cellular target recognition by HPV18 and HPV49 oncoproteins
To be published
|
|
6SIV
 
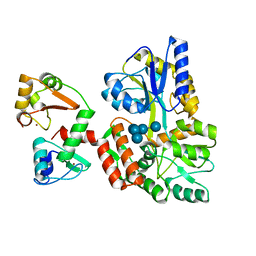 | | Structure of HPV16 E6 oncoprotein in complex with mutant IRF3 LxxLL motif | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | 著者 | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | 登録日 | 2019-08-12 | | 公開日 | 2019-08-21 | | 最終更新日 | 2022-03-30 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Deciphering the molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
2J8T
 
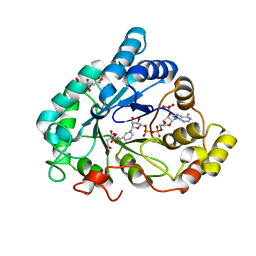 | | Human aldose reductase in complex with NADP and citrate at 0.82 angstrom | | 分子名称: | ALDO-KETO REDUCTASE FAMILY 1, MEMBER B1, CITRATE ANION, ... | | 著者 | Biadene, M, Hazemann, I, Cousido, A, Ginell, S, Sheldrick, G.M, Podjarny, A, Schneider, T.R. | | 登録日 | 2006-10-27 | | 公開日 | 2007-05-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (0.82 Å) | | 主引用文献 | The Atomic Resolution Structure of Human Aldose Reductase Reveals that Rearrangement of a Bound Ligand Allows the Opening of the Safety-Belt Loop.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5M2F
 
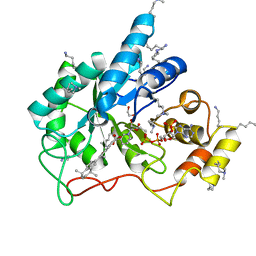 | | Crystal structure of human AKR1B10 complexed with NADP+ and the synthetic retinoid UVI2008 | | 分子名称: | 1,2-ETHANEDIOL, 3-bromo-4-[(1E)-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)prop-1-en-1-yl]benzoic acid, Aldo-keto reductase family 1 member B10, ... | | 著者 | Ruiz, F.X, Cousido-Siah, A, Mitschler, A, Porte, S, Alvarez, S, Dominguez, M, Alvarez, R, de Lera, A.R, Pares, X, Farres, J, Podjarny, A. | | 登録日 | 2016-10-12 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural basis for the inhibition of AKR1B10 by the C3 brominated TTNPB derivative UVI2008.
Chem. Biol. Interact., 276, 2017
|
|
1US0
 
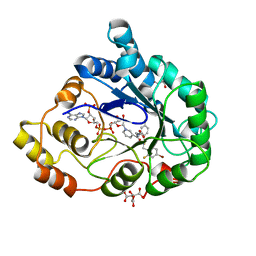 | | Human Aldose Reductase in complex with NADP+ and the inhibitor IDD594 at 0.66 Angstrom | | 分子名称: | ALDOSE REDUCTASE, CITRIC ACID, IDD594, ... | | 著者 | Howard, E.I, Sanishvili, R, Cachau, R.E, Mitschler, A, Chevrier, B, Barth, P, Lamour, V, Van Zandt, M, Sibley, E, Bon, C, Moras, D, Schneider, T.R, Joachimiak, A, Podjarny, A. | | 登録日 | 2003-11-16 | | 公開日 | 2004-05-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (0.66 Å) | | 主引用文献 | Ultrahigh Resolution Drug Design I: Details of Interactions in Human Aldose Reductase-Inhibitor Complex at 0.66 A.
Proteins, 55, 2004
|
|
3GHU
 
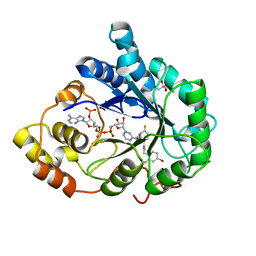 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Forth stage of radiation damage. | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | 登録日 | 2009-03-04 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3GHS
 
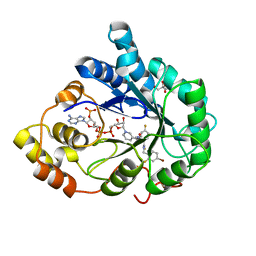 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Second stage of radiation damage. | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | 登録日 | 2009-03-04 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3GHT
 
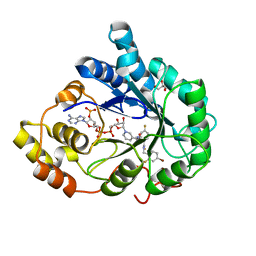 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Third stage of radiation damage. | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | 登録日 | 2009-03-04 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
