6Z1M
 
 | | Structure of an Ancestral glycosidase (family 1) bound to heme | | 分子名称: | 1,2-ETHANEDIOL, Ancestral reconstructed glycosidase, GLYCEROL, ... | | 著者 | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Oshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | 登録日 | 2020-05-14 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
6Z1H
 
 | | Ancestral glycosidase (family 1) | | 分子名称: | ANCESTRAL RECONSTRUCTED GLYCOSIDASE, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Hoshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | 登録日 | 2020-05-13 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
6TWW
 
 | | Variant W229D/F290W-19 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) | | 分子名称: | ACETATE ION, Beta-Lactamase (GNCA4), FORMIC ACID, ... | | 著者 | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L. | | 登録日 | 2020-01-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
6TXD
 
 | | Variant W229D/F290W-12 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) | | 分子名称: | ACETATE ION, Beta lactamase (GNCA4-12), FORMIC ACID, ... | | 著者 | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L. | | 登録日 | 2020-01-14 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
6TY6
 
 | | Variant W229D/F290W-2 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) bound to 5(6)-nitrobenzotriazole (TS-analog) | | 分子名称: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Beta lactamase (GNCA4-2), ... | | 著者 | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L, Ortega-Munoz, M, Santoyo-Gonzalez, F. | | 登録日 | 2020-01-15 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
6MS8
 
 | | Crystal Structure of Chalcone Isomerase from Medicago Truncatula Complexed with (2S) Naringenin | | 分子名称: | Chalcone-flavonone isomerase family protein, NARINGENIN | | 著者 | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | 登録日 | 2018-10-16 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6CJO
 
 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95S mutation. | | 分子名称: | Chalcone--flavonone isomerase 1, SULFATE ION | | 著者 | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | 登録日 | 2018-02-26 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6CJN
 
 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95T mutation | | 分子名称: | Chalcone--flavonone isomerase 1, SULFATE ION | | 著者 | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | 登録日 | 2018-02-26 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
3MOP
 
 | | The ternary Death Domain complex of MyD88, IRAK4, and IRAK2 | | 分子名称: | Interleukin-1 receptor-associated kinase 4, Interleukin-1 receptor-associated kinase-like 2, Myeloid differentiation primary response protein MyD88 | | 著者 | Lin, S.-C, Lo, Y.-C, Wu, H. | | 登録日 | 2010-04-23 | | 公開日 | 2010-06-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling.
Nature, 465, 2010
|
|
2QRA
 
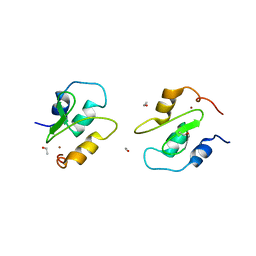 | | Crystal structure of XIAP BIR1 domain (P21 form) | | 分子名称: | Baculoviral IAP repeat-containing protein 4, ETHANOL, ZINC ION | | 著者 | Lin, S.-C. | | 登録日 | 2007-07-27 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the BIR1 domain of XIAP in two crystal forms
J.Mol.Biol., 372, 2007
|
|
8YD8
 
 | |
8YBX
 
 | |
8YD7
 
 | |
4YR8
 
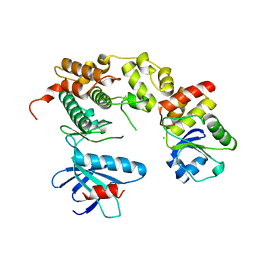 | | Crystal structure of JNK in complex with a regulator protein | | 分子名称: | CHLORIDE ION, Dual specificity protein phosphatase 16, Mitogen-activated protein kinase 8 | | 著者 | Liu, X, Wang, J, Wu, J.W, Wang, Z.X. | | 登録日 | 2015-03-14 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A conserved motif in JNK/p38-specific MAPK phosphatases as a determinant for JNK1 recognition and inactivation.
Nat Commun, 7, 2016
|
|
6KL5
 
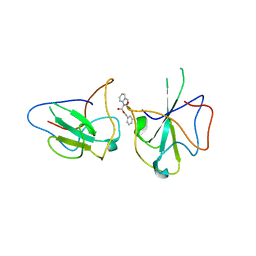 | | Structure of The N-terminal domain of Middle East respiratory syndrome coronavirus Nucleocapsid Protein complexed with Benzyl 2-(Hydroxymethyl)-1-Indolinecarboxylate | | 分子名称: | (phenylmethyl) (2S)-2-(hydroxymethyl)-2,3-dihydroindole-1-carboxylate, Nucleoprotein | | 著者 | Hou, M.H, Lin, S.M, Hsu, J.N, Wang, Y.S. | | 登録日 | 2019-07-29 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL6
 
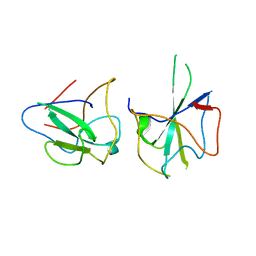 | | Crystal structure of MERS-CoV N-NTD complexed with 5-Benzyloxygramine | | 分子名称: | N,N-dimethyl-1-(5-phenylmethoxy-1H-indol-3-yl)methanamine, Nucleoprotein | | 著者 | Hou, M.H, Lin, S.M, Wang, Y.S, Hsu, J.N. | | 登録日 | 2019-07-29 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.
J.Med.Chem., 63, 2020
|
|
6KL2
 
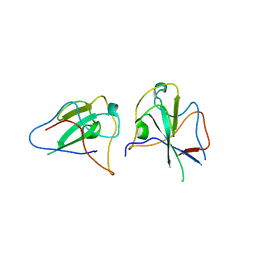 | |
4UHB
 
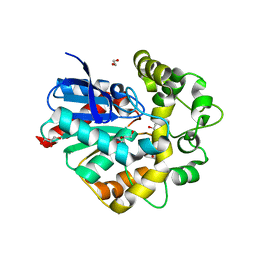 | | Laboratory evolved variant R-C1 of potato epoxide hydrolase StEH1 | | 分子名称: | 1,2-ETHANEDIOL, EPOXIDE HYDROLASE, GLYCEROL | | 著者 | Nilsson, M.T.I, Carlsson, A.J, Dobritzsch, D, Widersten, M. | | 登録日 | 2015-03-23 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Laboratory Evolved Enzymes Provide Snapshots of the Development of Enantioconvergence in Enzyme-Catalyzed Epoxide Hydrolysis.
Chembiochem, 17, 2016
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | 分子名称: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | 著者 | Hong, W, Chen, H, Wu, Q, Lin, T. | | 登録日 | 2019-09-23 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.45 Å) | | 主引用文献 | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3HR7
 
 | |
3QT0
 
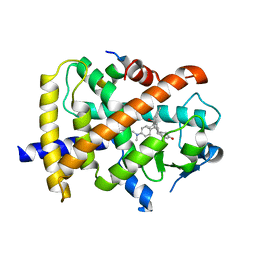 | | Revealing a steroid receptor ligand as a unique PPARgamma agonist | | 分子名称: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, Nuclear receptor coactivator 1 peptide, Peroxisome proliferator-activated receptor gamma | | 著者 | Rong, H. | | 登録日 | 2011-02-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.496 Å) | | 主引用文献 | Revealing a steroid receptor ligand as a unique PPAR gamma agonist.
Cell Res., 22, 2012
|
|
8IV3
 
 | |
8J6X
 
 | |
8IQJ
 
 | | Crystal structure of SARS-CoV2 N-NTD | | 分子名称: | Nucleoprotein | | 著者 | Hong, J.Y, Hou, M.H. | | 登録日 | 2023-03-16 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Targeting protein-protein interaction interfaces with antiviral N protein inhibitor in SARS-CoV-2.
Biophys.J., 123, 2024
|
|
7F63
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45 (Focused refinement of S-RBD and chAb-45 region) | | 分子名称: | RBD-chAb45, Heavy chain, Light chain, ... | | 著者 | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | 登録日 | 2021-06-24 | | 公開日 | 2021-08-04 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure-guided antibody cocktail for prevention and treatment of COVID-19.
Plos Pathog., 17, 2021
|
|
