3FPD
 
 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | 分子名称: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | 著者 | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | 登録日 | 2009-01-05 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5V21
 
 | |
1T8R
 
 | |
1T8Y
 
 | |
9C11
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36R at cryogenic temperature | | 分子名称: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | 著者 | Zhang, Y, Schlessman, J.L, Siegler, M.A, Garcia-Moreno E, B. | | 登録日 | 2024-05-28 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Domain-swapping promoted by the introduction of a charge in the hydrophobic interior of a protein
To Be Published
|
|
5UQ7
 
 | | 70S ribosome complex with dnaX mRNA stemloop and E-site tRNA ("in" conformation) | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | 登録日 | 2017-02-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
6J1L
 
 | | Crystal Structure Analysis of the ROR gamma(C455E) | | 分子名称: | 2-[4-(ethylsulfonyl)phenyl]-N-[2'-fluoro-4'-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)[1,1'-biphenyl]-4-yl]acetamide, Nuclear receptor ROR-gamma | | 著者 | zhang, Y, Li, C.C, wu, X.S. | | 登録日 | 2018-12-28 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery and Characterization of XY101, a Potent, Selective, and Orally Bioavailable ROR gamma Inverse Agonist for Treatment of Castration-Resistant Prostate Cancer.
J.Med.Chem., 62, 2019
|
|
6CNF
 
 | | Yeast RNA polymerase III elongation complex | | 分子名称: | DNA (79-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2018-03-08 | | 公開日 | 2018-08-22 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNB
 
 | | Yeast RNA polymerase III initial transcribing complex | | 分子名称: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2018-03-08 | | 公開日 | 2018-08-22 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNC
 
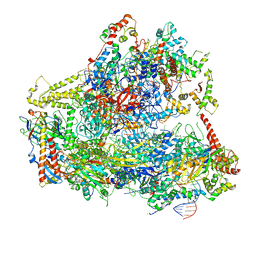 | | Yeast RNA polymerase III open complex | | 分子名称: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2018-03-08 | | 公開日 | 2018-08-22 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CND
 
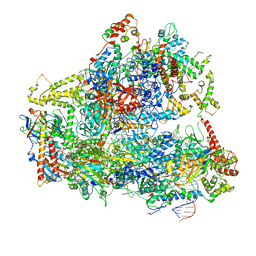 | | Yeast RNA polymerase III natural open complex (nOC) | | 分子名称: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2018-03-08 | | 公開日 | 2018-08-22 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
5W66
 
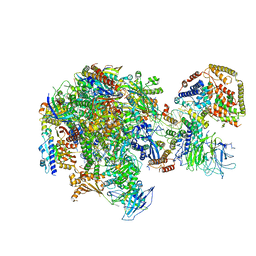 | | RNA polymerase I Initial Transcribing Complex State 3 | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2017-06-16 | | 公開日 | 2017-07-26 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W65
 
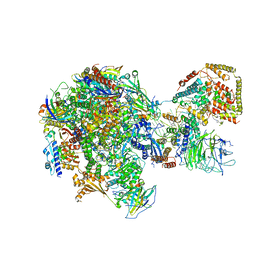 | | RNA polymerase I Initial Transcribing Complex State 2 | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2017-06-16 | | 公開日 | 2017-08-02 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W64
 
 | | RNA Polymerase I Initial Transcribing Complex State 1 | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2017-06-16 | | 公開日 | 2017-07-26 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W5Y
 
 | | RNA polymerase I Initial Transcribing Complex | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Han, Y, He, Y. | | 登録日 | 2017-06-16 | | 公開日 | 2017-07-26 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
8IHM
 
 | | Eaf3 CHD domain bound to the nucleosome | | 分子名称: | Chromatin modification-related protein EAF3, DNA (164-MER), DNA (165-MER), ... | | 著者 | Zhang, Y, Gang, C. | | 登録日 | 2023-02-23 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHN
 
 | | Cryo-EM structure of the Rpd3S core complex | | 分子名称: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | 著者 | Zhang, Y, Gang, C. | | 登録日 | 2023-02-23 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHT
 
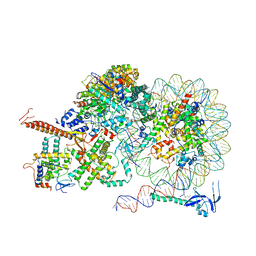 | | Rpd3S bound to the nucleosome | | 分子名称: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | 著者 | Zhang, Y, Gang, C. | | 登録日 | 2023-02-23 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
7K65
 
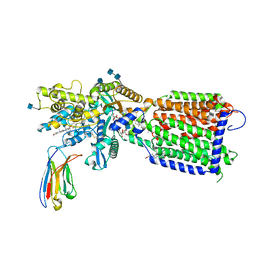 | | Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | 著者 | Zhang, Y, Bulkley, D.P, Liang, J, Manglik, A, Cheng, Y, Beachy, P.A. | | 登録日 | 2020-09-18 | | 公開日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Hedgehog pathway activation through nanobody-mediated conformational blockade of the Patched sterol conduit.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4G8L
 
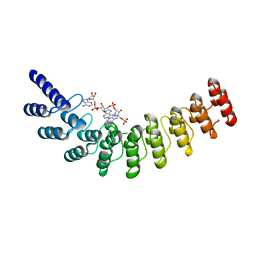 | | Active state of intact sensor domain of human RNase L with 2-5A bound | | 分子名称: | 2-5A-dependent ribonuclease, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE | | 著者 | Han, Y, Whitney, G, Donovan, J, Korennykh, A. | | 登録日 | 2012-07-23 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Innate Immune Messenger 2-5A Tethers Human RNase L into Active High-Order Complexes.
Cell Rep, 2, 2012
|
|
4G8K
 
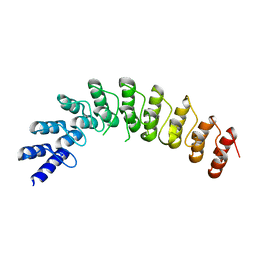 | |
4OAV
 
 | | Complete human RNase L in complex with 2-5A (5'-ppp heptamer), AMPPCP and RNA substrate. | | 分子名称: | (2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-hydroxy-2-({[(S)-hydroxy{[(2R,3S,4S)-4-hydroxy-2-(hydroxymethyl)tetrahydrofuran-3-yl]oxy}phosphoryl]oxy}methyl)tetrahydrofuran-3-yl dihydrogen phosphate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | 著者 | Han, Y, Donovan, J, Rath, S, Whitney, G, Chitrakar, A, Korennykh, A. | | 登録日 | 2014-01-06 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response.
Science, 343, 2014
|
|
4OAU
 
 | | Complete human RNase L in complex with biological activators. | | 分子名称: | 2-5A-dependent ribonuclease, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Han, Y, Donovan, J, Rath, S, Whitney, G, Chitrakar, A, Korennykh, A. | | 登録日 | 2014-01-06 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response.
Science, 343, 2014
|
|
1O7Q
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | 分子名称: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | 著者 | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | 登録日 | 2002-11-12 | | 公開日 | 2003-11-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
1O7O
 
 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | 分子名称: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | 登録日 | 2002-11-11 | | 公開日 | 2003-11-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity.
Biochemistry, 42, 2003
|
|
