7S14
 
 | | Crystal structure of putative NAD(P)H-flavin oxidoreductase from Haemophilus influenzae 86-028NP | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Kim, Y, Maltseva, N, Endres, M, Crofts, T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-08-31 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Functional and Structural Characterization of Diverse NfsB Chloramphenicol Reductase Enzymes from Human Pathogens.
Microbiol Spectr, 10, 2022
|
|
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | 分子名称: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | 著者 | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | 登録日 | 2017-11-07 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
2VRC
 
 | | Crystal structure of the Citrobacter sp. triphenylmethane reductase complexed with NADP(H) | | 分子名称: | TRIPHENYLMETHANE REDUCTASE | | 著者 | Kim, Y, Park, H.J, Kwak, S.N, Lee, J.S, Oh, T.K, Kim, M.H. | | 登録日 | 2008-03-31 | | 公開日 | 2008-09-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase.
J.Biol.Chem., 283, 2008
|
|
2VRB
 
 | | Crystal structure of the Citrobacter sp. triphenylmethane reductase complexed with NADP(H) | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIPHENYLMETHANE REDUCTASE | | 著者 | Kim, Y, Park, H.J, Kwak, S.N, Lee, J.S, Oh, T.K, Kim, M.H. | | 登録日 | 2008-03-31 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase.
J.Biol.Chem., 283, 2008
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | 分子名称: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | 著者 | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2013-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
1ACP
 
 | |
2LA2
 
 | |
7CAZ
 
 | |
7CAW
 
 | |
7CAX
 
 | | Crystal structure of bacterial reductase | | 分子名称: | 1,2-ETHANEDIOL, 3-oxoacyl-ACP reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Kim, Y, Lee, W.C. | | 登録日 | 2020-06-10 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.846 Å) | | 主引用文献 | Crystal structure of bacterial reductase
To be published
|
|
2JL1
 
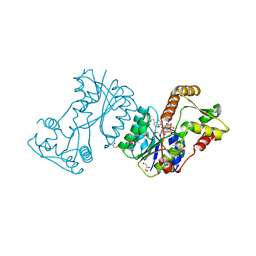 | | Structural insight into bioremediation of triphenylmethane dyes by Citrobacter sp. triphenylmethane reductase | | 分子名称: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIPHENYLMETHANE REDUCTASE | | 著者 | Kim, Y, Park, H.J, Kwak, S.N, Kim, M.H. | | 登録日 | 2008-09-02 | | 公開日 | 2008-09-23 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase
J.Biol.Chem., 283, 2008
|
|
6PU9
 
 | | Crystal Structure of the Type B Chloramphenicol O-Acetyltransferase from Vibrio vulnificus | | 分子名称: | 1,2-ETHANEDIOL, Acetyltransferase, CHLORIDE ION | | 著者 | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-07-17 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
6PUA
 
 | | The 2.0 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-07-18 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
1ERI
 
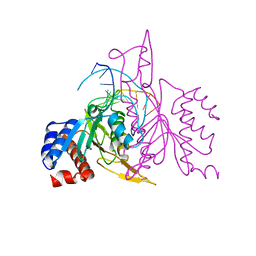 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | 分子名称: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | 著者 | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | 登録日 | 1994-05-18 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
4DCD
 
 | | 1.6A resolution structure of PolioVirus 3C Protease Containing a covalently bound dipeptidyl inhibitor | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Genome polyprotein, ... | | 著者 | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.-C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | 登録日 | 2012-01-17 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
4F49
 
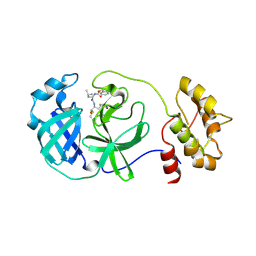 | | 2.25A resolution structure of Transmissible Gastroenteritis Virus Protease containing a covalently bound Dipeptidyl Inhibitor | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | 著者 | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.-C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | 登録日 | 2012-05-10 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
6X4I
 
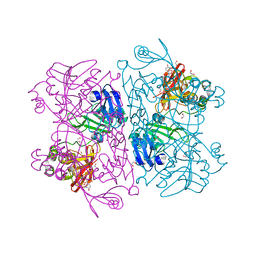 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate | | 分子名称: | 1,2-ETHANEDIOL, 3'-URIDINEMONOPHOSPHATE, SODIUM ION, ... | | 著者 | Chang, C, Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-22 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
4ZQN
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor P41 | | 分子名称: | 2-chloro-N,N-dimethyl-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]benzamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZQM
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with XMP and NAD | | 分子名称: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZQP
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor MAD1 | | 分子名称: | 5'-O-({1-[(2E)-4-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1,3-dihydro-2-benzofuran-5-yl)-2-methylbut-2-en-1-yl]-1H-1,2,3-triazol-4-yl}methyl)adenosine, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZQR
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis | | 分子名称: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-11 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.692 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
4ZQO
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with IMP and the inhibitor Q67 | | 分子名称: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | 著者 | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-10 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
5HPF
 
 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain from Acinetobacter | | 分子名称: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | 登録日 | 2016-01-20 | | 公開日 | 2016-09-07 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.309 Å) | | 主引用文献 | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
5HPI
 
 | | Crystal Structure of the Double Mutant of PobR Transcription Factor Inducer Binding Domain-3-Hydroxy Benzoic Acid complex from Acinetobacter | | 分子名称: | 1,2-ETHANEDIOL, 3-HYDROXYBENZOIC ACID, SULFATE ION, ... | | 著者 | Kim, Y, Tesar, C, Jedrejczak, R, Jha, R, Strauss, C.E.M, Joachimiak, A. | | 登録日 | 2016-01-20 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.963 Å) | | 主引用文献 | A microbial sensor for organophosphate hydrolysis exploiting an engineered specificity switch in a transcription factor.
Nucleic Acids Res., 44, 2016
|
|
6JHW
 
 | | Structure of anti-CRISPR AcrIIC3 and NmeCas9 HNH | | 分子名称: | AcrIIC3, CRISPR-associated endonuclease Cas9 | | 著者 | Suh, J.Y, Lee, B.J, Lee, S.J, Kim, Y. | | 登録日 | 2019-02-19 | | 公開日 | 2019-08-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Anti-CRISPR AcrIIC3 discriminates between Cas9 orthologs via targeting the variable surface of the HNH nuclease domain.
Febs J., 286, 2019
|
|
