5QII
 
 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-(3-(1-(4-CHLOROPHENYL)CYCLOPROPYL) -[1,2,4]TRIAZOLO[4,3-A]PYRIDIN-8-YL)PROPAN-2-OL | | 分子名称: | 2-{3-[1-(4-chlorophenyl)cyclopropyl][1,2,4]triazolo[4,3-a]pyridin-8-yl}propan-2-ol, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Sheriff, S. | | 登録日 | 2018-07-03 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Discovery of Clinical Candidate BMS-823778 as an Inhibitor of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 9, 2018
|
|
5QIJ
 
 | | CRYSTAL STRUCTURE OF MURINE 11B- HYDROXYSTEROIDDEHYDROGENASE COMPLEXED WITH 2-(3-(1-(4- CHLOROPHENYL)CYCLOPROPYL)-[1,2,4]TRIAZOLO[4,3-A]PYRIDIN-8- YL)PROPAN-2-OL | | 分子名称: | 2-{3-[1-(4-chlorophenyl)cyclopropyl][1,2,4]triazolo[4,3-a]pyridin-8-yl}propan-2-ol, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Sheriff, S. | | 登録日 | 2018-07-03 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Discovery of Clinical Candidate BMS-823778 as an Inhibitor of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 9, 2018
|
|
8RI9
 
 | | Late alpha-Synuclein fibril structure from liquid-liquid phase separations. | | 分子名称: | Alpha-synuclein | | 著者 | De Simone, A, Barritt, J.D, Chen, S, Cascella, R, Cecchi, C, Bigi, A, Jarvis, J.A, Chiti, F, Dobson, C.M, Fusco, G. | | 登録日 | 2023-12-18 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure-Toxicity Relationship in Intermediate Fibrils from alpha-Synuclein Condensates.
J.Am.Chem.Soc., 146, 2024
|
|
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | 著者 | Zhang, H, Luo, C. | | 登録日 | 2019-09-09 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.981 Å) | | 主引用文献 | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6KX2
 
 | | Crystal structure of GDP bound RhoA protein | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | 著者 | Zhang, H, Luo, C. | | 登録日 | 2019-09-09 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.454 Å) | | 主引用文献 | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
8YBF
 
 | | Crystal structure of canine distemper virus hemagglutinin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein | | 著者 | Fukuhara, H, Yumoto, K, Sako, M, Kajikawa, M, Ose, T, Hashiguchi, T, Kamishikiryo, J, Maita, N, Kuroki, K, Maenaka, K. | | 登録日 | 2024-02-13 | | 公開日 | 2024-07-31 | | 最終更新日 | 2024-08-14 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Glycan-shielded homodimer structure and dynamical features of the canine distemper virus hemagglutinin relevant for viral entry and efficient vaccination.
Elife, 12, 2024
|
|
6IEG
 
 | | Crystal structure of human MTR4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Exosome RNA helicase MTR4, MAGNESIUM ION | | 著者 | Chen, J.Y, Yun, C.H. | | 登録日 | 2018-09-14 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.55 Å) | | 主引用文献 | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
6IEH
 
 | | Crystal structures of the hMTR4-NRDE2 complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | 著者 | Chen, J.Y, Yun, C.H. | | 登録日 | 2018-09-14 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.892 Å) | | 主引用文献 | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
3BY7
 
 | |
6KTC
 
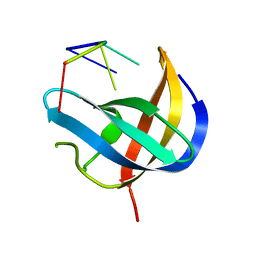 | | Crystal structure of YBX1 CSD with m5C RNA | | 分子名称: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*GP*(5MC)P*CP*U)-3') | | 著者 | Zou, F, Li, S. | | 登録日 | 2019-08-27 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.008 Å) | | 主引用文献 | DrosophilaYBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KUG
 
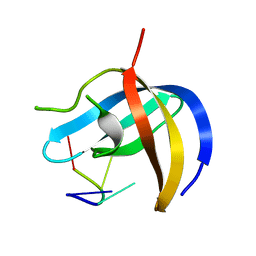 | | Crystal structure of YBX1 CSD with RNA | | 分子名称: | Nuclease-sensitive element-binding protein 1, RNA (5'-R(P*GP*CP*CP*U)-3') | | 著者 | Zou, F, Li, S. | | 登録日 | 2019-09-02 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | DrosophilaYBX1 homolog YPS promotes ovarian germ line stem cell development by preferentially recognizing 5-methylcytosine RNAs.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4XIY
 
 | | Crystal structure of ketol-acid reductoisomerase from Azotobacter | | 分子名称: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Ketol-acid reductoisomerase, ... | | 著者 | Spatzal, T, Cahn, J.K.B, Wiig, J.A, Einsle, O, Hu, Y, Ribbe, M.W, Arnold, F.H. | | 登録日 | 2015-01-07 | | 公開日 | 2015-04-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
3KDU
 
 | |
3KDT
 
 | |
7CN0
 
 | | Cryo-EM structure of K+-bound hERG channel | | 分子名称: | POTASSIUM ION, potassium channel 1 | | 著者 | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | 登録日 | 2020-07-29 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7CN1
 
 | | Cryo-EM structure of K+-bound hERG channel in the presence of astemizole | | 分子名称: | POTASSIUM ION, potassium channel | | 著者 | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | 登録日 | 2020-07-29 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
5NP7
 
 | |
7N6W
 
 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U2 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | 分子名称: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, S, Wang, K, Mao, Y. | | 登録日 | 2021-06-09 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-12-08 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
7N6U
 
 | | Structure of uncleaved HIV-1 JR-FL Env glycoprotein trimer in state U1 bound to small Molecule HIV-1 Entry Inhibitor BMS-378806 | | 分子名称: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, S, Wang, K, Mao, Y. | | 登録日 | 2021-06-09 | | 公開日 | 2021-09-22 | | 最終更新日 | 2021-12-08 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Asymmetric Structures and Conformational Plasticity of the Uncleaved Full-Length Human Immunodeficiency Virus Envelope Glycoprotein Trimer.
J.Virol., 95, 2021
|
|
5IXJ
 
 | |
3C0F
 
 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | 分子名称: | Uncharacterized protein AF_1514 | | 著者 | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | 登録日 | 2008-01-20 | | 公開日 | 2008-02-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
7C6Q
 
 | | Novel natural PPARalpha agonist with a unique binding mode | | 分子名称: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Peroxisome proliferator-activated receptor alpha | | 著者 | Tian, S.Y, Wang, R, Zheng, W.L, Li, Y. | | 登録日 | 2020-05-22 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structural Basis for PPARs Activation by The Dual PPAR alpha / gamma Agonist Sanguinarine: A Unique Mode of Ligand Recognition.
Molecules, 26, 2021
|
|
1AV8
 
 | | RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT FROM E. COLI | | 分子名称: | MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | 著者 | Han, S, Arvai, A, Tainer, J.A. | | 登録日 | 1997-09-30 | | 公開日 | 1998-10-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
6E66
 
 | |
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | 分子名称: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | 著者 | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | 登録日 | 2015-09-12 | | 公開日 | 2016-03-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
