5LXD
 
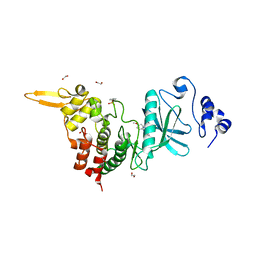 | | Crystal structure of DYRK2 in complex with EHT 1610 (compound 2) | | 分子名称: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2-fluoranyl-4-methoxy-phenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | 著者 | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2016-09-20 | | 公開日 | 2016-10-26 | | 最終更新日 | 2017-01-11 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
5LXC
 
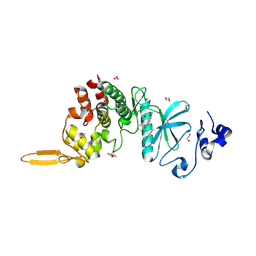 | | Crystal structure of DYRK2 in complex with EHT 5372 (Compound 1) | | 分子名称: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 2, methyl 9-[(2,4-dichlorophenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | 著者 | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Besson, T, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2016-09-20 | | 公開日 | 2016-10-26 | | 最終更新日 | 2017-01-11 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | An Unusual Binding Model of the Methyl 9-Anilinothiazolo[5,4-f] quinazoline-2-carbimidates (EHT 1610 and EHT 5372) Confers High Selectivity for Dual-Specificity Tyrosine Phosphorylation-Regulated Kinases.
J. Med. Chem., 59, 2016
|
|
7SSE
 
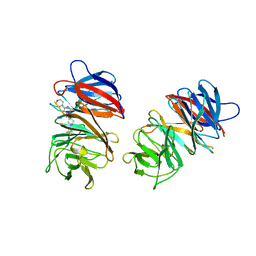 | | Crystal structure of the WDR domain of human DCAF1 in complex with CYCA-117-70 | | 分子名称: | DDB1- and CUL4-associated factor 1, N-[(3R)-1-(3-fluorophenyl)piperidin-3-yl]-6-(morpholin-4-yl)pyrimidin-4-amine | | 著者 | Kimani, S, Owen, J, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Shahani, V.M, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2021-11-10 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery of a Novel DCAF1 Ligand Using a Drug-Target Interaction Prediction Model: Generalizing Machine Learning to New Drug Targets.
J.Chem.Inf.Model., 63, 2023
|
|
5LEV
 
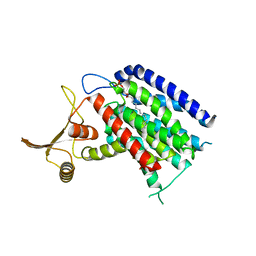 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) | | 分子名称: | UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, UNKNOWN LIGAND | | 著者 | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2016-06-30 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
5LWN
 
 | | Crystal structure of JAK3 in complex with Compound 5 (FM409) | | 分子名称: | (2~{S})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]propanamide, (~{Z})-2-cyano-~{N},~{N}-dimethyl-3-[5-[3-[(1~{S},2~{R})-2-methylcyclohexyl]-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1,4,6,8,11-pentaen-4-yl]furan-2-yl]prop-2-enamide, 1,2-ETHANEDIOL, ... | | 著者 | Chaikuad, A, Forster, M, Mukhopadhyay, S, Kupinska, K, Ellis, K, Mahajan, P, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2016-09-18 | | 公開日 | 2016-10-26 | | 最終更新日 | 2016-11-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Selective JAK3 Inhibitors with a Covalent Reversible Binding Mode Targeting a New Induced Fit Binding Pocket.
Cell Chem Biol, 23, 2016
|
|
3NHV
 
 | | Crystal Structure of BH2092 protein from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR228F | | 分子名称: | BH2092 protein, CHLORIDE ION | | 著者 | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-06-14 | | 公開日 | 2010-08-11 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target BhR228F
To be Published, 2010
|
|
6T6A
 
 | | Crystal structure of DYRK1A complexed with KuFal319 (compound 11) | | 分子名称: | 4-chloranyl-5~{H}-cyclohepta[b]indol-10-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | 著者 | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-10-18 | | 公開日 | 2019-12-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | [ b ]-Annulated Halogen-Substituted Indoles as Potential DYRK1A Inhibitors.
Molecules, 24, 2019
|
|
1JJG
 
 | | Solution Structure of Myxoma Virus Protein M156R | | 分子名称: | M156R | | 著者 | Ramelot, T.A, Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2001-07-05 | | 公開日 | 2002-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha.
J.Mol.Biol., 322, 2002
|
|
3NRO
 
 | | Crystal Structure of putative transcriptional factor Lmo1026 from Listeria monocytogenes (FRAGMENT 52-321), Northeast Structural Genomics Consortium Target LmR194 | | 分子名称: | Lmo1026 protein | | 著者 | Kuzin, A, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-06-30 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target LmR194
To be Published
|
|
3NRN
 
 | | Crystal Structure of PF1083 protein from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR223 | | 分子名称: | ADENOSINE MONOPHOSPHATE, uncharacterized protein PF1083 | | 著者 | Seetharaman, J, Su, M, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-06-30 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target PfR223
To be Published
|
|
5LF8
 
 | | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17) | | 分子名称: | ETHYL MERCURY ION, Nucleoside diphosphate-linked moiety X motif 17, PHOSPHATE ION | | 著者 | Mathea, S, Tallant, C, Salah, E, Wang, D, Velupillai, S, Nowak, R, Oerum, S, Krojer, T, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | 登録日 | 2016-06-30 | | 公開日 | 2017-08-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17)
To Be Published
|
|
3EW7
 
 | | Crystal structure of the Lmo0794 protein from Listeria monocytogenes. Northeast Structural Genomics Consortium target LmR162. | | 分子名称: | Lmo0794 protein | | 著者 | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Wang, D, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-10-14 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Crystal structure of the Lmo0794 protein from Listeria monocytogenes.
To be Published
|
|
3EAP
 
 | | Crystal structure of the RhoGAP domain of ARHGAP11A | | 分子名称: | Rho GTPase-activating protein 11A, UNKNOWN ATOM OR ION | | 著者 | Shen, Y, Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | 登録日 | 2008-08-26 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of the RhoGAP domain of ARHGAP11A
To be Published
|
|
3NGW
 
 | | Crystal Structure of Molybdopterin-guanine dinucleotide biosynthesis protein A from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR189 | | 分子名称: | CALCIUM ION, Molybdopterin-guanine dinucleotide biosynthesis protein A (MobA) | | 著者 | Forouhar, F, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-06-13 | | 公開日 | 2010-08-11 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Northeast Structural Genomics Consortium Target GR189
To be Published
|
|
3NZL
 
 | | Crystal Structure of the N-terminal domain of DNA-binding protein SATB1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4435B | | 分子名称: | DNA-binding protein SATB1 | | 著者 | Forouhar, F, Abashidze, M, Seetharaman, J, Kuzin, A.P, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-07-16 | | 公開日 | 2010-09-22 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.199 Å) | | 主引用文献 | Crystal Structure of the N-terminal domain of DNA-binding protein SATB1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4435B (CASP Target)
To be Published
|
|
3FIF
 
 | | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A. | | 分子名称: | Uncharacterized ligand, Uncharacterized lipoprotein ygdR | | 著者 | Kuzin, A.P, Su, M, Seetharaman, J, Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-12-11 | | 公開日 | 2009-01-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the ygdR protein from E.coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
3FOK
 
 | | Crystal Structure of Cgl0159 From Corynebacterium glutamicum (Brevibacterium flavum). Northeast Structural Genomics Target CgR115 | | 分子名称: | uncharacterized protein Cgl0159 | | 著者 | Seetharaman, J, Neely, H, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-12-30 | | 公開日 | 2009-01-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Cgl0159 From Corynebacterium glutamicum (Brevibacterium flavum). Northeast Structural Genomics Target CgR115
To be Published
|
|
1YEZ
 
 | | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30. | | 分子名称: | MM1357 | | 著者 | Rossi, P, Aramini, J.M, Swapna, G.V.T, Huang, Y.P, Xiao, R, Ho, C.K, Ma, L.C, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-12-29 | | 公開日 | 2005-02-22 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30.
To be Published
|
|
3EQE
 
 | | Crystal structure of the YubC protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR112. | | 分子名称: | FE (III) ION, Putative cystein dioxygenase | | 著者 | Vorobiev, S.M, Su, M, Seetharaman, J, Benach, J, Forouhar, F, Clayton, G, Cooper, B, Wang, H, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-09-30 | | 公開日 | 2008-10-14 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Crystal structure of the YubC protein from Bacillus subtilis.
To be Published
|
|
3ESH
 
 | | Crystal structure of a probable metal-dependent hydrolase from Staphylococcus aureus. Northeast Structural Genomics target ZR314 | | 分子名称: | Protein similar to metal-dependent hydrolase | | 著者 | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-10-06 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a probable metal-dependent hydrolase from
Staphylococcus aureus. Northeast Structural Genomics target ZR314
To be Published
|
|
1YDM
 
 | | X-Ray structure of Northeast Structural Genomics target SR44 | | 分子名称: | Hypothetical protein yqgN, SULFATE ION | | 著者 | Kuzin, A.P, Jianwei, S, Vorobiev, S, Acton, T, Xia, R, Ma, L.-C, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-12-24 | | 公開日 | 2005-01-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-Ray structure of Northeast Structural Genomics target SR44
To be Published
|
|
9BHS
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-9939 compound | | 分子名称: | (4P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrole-3-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN LIGAND | | 著者 | kimani, S, Dong, A, Li, Y, Seitova, A, Al-Awar, R, Wilson, B, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2024-04-21 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-9939 compound
To be published
|
|
9BHR
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-40155 compound | | 分子名称: | (4P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-{(1E)-3-[(2-methoxyethyl)amino]-3-oxoprop-1-en-1-yl}-1H-pyrrole-3-carboxamide, DDB1- and CUL4-associated factor 1 | | 著者 | kimani, S, Dong, A, Li, Y, Seitova, A, Al-Awar, R, Krausser, C, Wilson, B, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | 登録日 | 2024-04-21 | | 公開日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-40155 compound
To be published
|
|
1Y0H
 
 | | Structure of Rv0793 from Mycobacterium tuberculosis | | 分子名称: | ACETATE ION, hypothetical protein Rv0793 | | 著者 | Lemieux, M.J, Ference, C, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2004-11-15 | | 公開日 | 2004-12-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of Rv0793, a hypothetical monooxygenase from M. tuberculosis
J.STRUCT.FUNCT.GENOM., 6, 2005
|
|
1KPH
 
 | | Crystal Structure of mycolic acid cyclopropane synthase CmaA1 complexed with SAH and DDDMAB | | 分子名称: | CARBONATE ION, CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 1, DIDECYL-DIMETHYL-AMMONIUM, ... | | 著者 | Huang, C.-C, Smith, C.V, Jacobs Jr, W.R, Glickman, M.S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2001-12-30 | | 公開日 | 2002-01-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis
J.Biol.Chem., 277, 2002
|
|
