4DAM
 
 | | Crystal structure of small single-stranded DNA-binding protein from Streptomyces coelicolor | | 分子名称: | Single-stranded DNA-binding protein 1 | | 著者 | Filic, Z, Herron, P, Ivic, N, Luic, M, Manjasetty, B.A, Paradzik, T, Vujaklija, D. | | 登録日 | 2012-01-13 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-function relationships of two paralogous single-stranded DNA-binding proteins from Streptomyces coelicolor: implication of SsbB in chromosome segregation during sporulation.
Nucleic Acids Res., 41, 2013
|
|
2WZE
 
 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | 分子名称: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, GLYCEROL, ... | | 著者 | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | 登録日 | 2009-11-27 | | 公開日 | 2010-08-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2WYS
 
 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | 分子名称: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, PHOSPHATE ION, ... | | 著者 | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | 登録日 | 2009-11-20 | | 公開日 | 2010-08-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
3FW0
 
 | | Structure of Peptidyl-alpha-hydroxyglycine alpha-Amidating Lyase (PAL) bound to alpha-hydroxyhippuric acid (non-peptidic substrate) | | 分子名称: | (2S)-hydroxy[(phenylcarbonyl)amino]ethanoic acid, CALCIUM ION, MERCURY (II) ION, ... | | 著者 | Chufan, E.E, De, M, Eipper, B.A, Mains, R.E, Amzel, L.M. | | 登録日 | 2009-01-16 | | 公開日 | 2009-09-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme.
Structure, 17, 2009
|
|
3DBR
 
 | |
3DPL
 
 | |
3FVZ
 
 | | Structure of Peptidyl-alpha-hydroxyglycine alpha-Amidating Lyase (PAL) | | 分子名称: | ACETATE ION, CALCIUM ION, FE (III) ION, ... | | 著者 | Chufan, E.E, De, M, Eipper, B.A, Mains, R.E, Amzel, L.M. | | 登録日 | 2009-01-16 | | 公開日 | 2009-09-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme.
Structure, 17, 2009
|
|
4BZ0
 
 | | Structural characterization using Sulfur-SAD of the cytoplasmic domain of Burkholderia pseudomallei PilO2Bp, an actin-like protein component of a Type IVb R64-derivative pilus machinery. | | 分子名称: | POTASSIUM ION, PUTATIVE TYPE IV PILUS BIOSYNTHESIS PROTEIN | | 著者 | Lassaux, P, Manjasetty, B.A, Conchillo-Sole, O, Yero, D, Gourlay, L, Perletti, L, Daura, X, Belrhali, H, Bolognesi, M. | | 登録日 | 2013-07-22 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Redefining the Pf06864 Pfam Family Based on Burkholderia Pseudomallei Pilo2BP S-Sad Crystal Structure.
Plos One, 9, 2014
|
|
2XH6
 
 | | Clostridium perfringens enterotoxin | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, HEAT-LABILE ENTEROTOXIN B CHAIN, octyl beta-D-glucopyranoside | | 著者 | Briggs, D.C, Naylor, C.E, Smedley III, J.G, MCClane, B.A, Basak, A.K. | | 登録日 | 2010-06-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
4CQ6
 
 | | The crystal structure of the allene oxide cyclase 2 from Arabidopsis thaliana with bound inhibitor - vernolic acid | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (9Z)-11-[(2R,3S)-3-pentyloxiran-2-yl]undec-9-enoic acid, ALLENE OXIDE CYCLASE 2, ... | | 著者 | Terlecka, B.A, Pollmann, S, Hofmann, E. | | 登録日 | 2014-02-11 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Ligand-Bound Structures of the Aoc2 from A. Thaliana and the Implications for the Catalytic Mechanism
To be Published
|
|
4CQ7
 
 | | The crystal structure of the allene oxide cyclase 2 from Arabidopsis thaliana with bound product - OPDA | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 9S,13R-12-OXOPHYTODIENOIC ACID, ALLENE OXIDE CYCLASE 2, ... | | 著者 | Terlecka, B.A, Pollmann, S, Hofmann, E. | | 登録日 | 2014-02-11 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Ligand-Bound Structures of the Aoc2 from A. Thaliana and the Implications for the Catalytic Mechanism
To be Published
|
|
3DQV
 
 | | Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation | | 分子名称: | Cullin-5, NEDD8, Rbx1, ... | | 著者 | Duda, D.M, Borg, L.A, Scott, D.C, Hunt, H.W, Hammel, M, Schulman, B.A. | | 登録日 | 2008-07-09 | | 公開日 | 2008-09-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.
Cell(Cambridge,Mass.), 134, 2008
|
|
3EA1
 
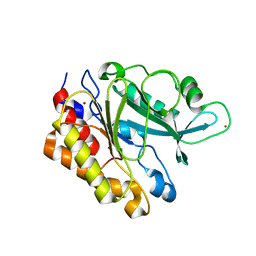 | | Crystal Structure of the Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | 分子名称: | 1-phosphatidylinositol phosphodiesterase, ZINC ION | | 著者 | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | 登録日 | 2008-08-24 | | 公開日 | 2009-04-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
4EE2
 
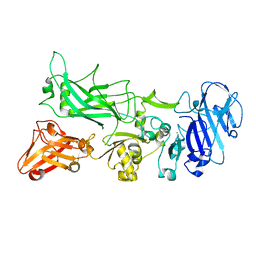 | |
4EOU
 
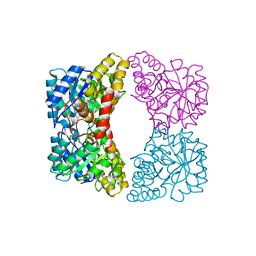 | |
3DBH
 
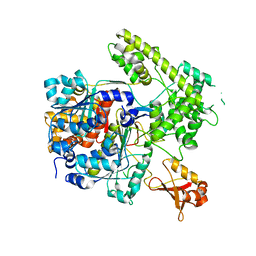 | |
3DD2
 
 | |
8R5H
 
 | | Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2 | | 分子名称: | (2~{S},4~{R})-1-[(2~{S})-2-[2-[2-[2-[2-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]ethoxy]ethoxy]ethoxy]ethanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-2,3-dihydro-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 5-azanylpentan-2-one, Bromodomain-containing protein 4, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-11-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
8RHZ
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | 分子名称: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | 著者 | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2023-12-17 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 2024
|
|
8Q7H
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | 分子名称: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | 著者 | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | 登録日 | 2023-08-16 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 2024
|
|
8Q7E
 
 | |
8U0X
 
 | | Yeast Pex6 N1(1-184) Domain | | 分子名称: | Peroxisomal ATPase PEX6 | | 著者 | Gardner, B.M, Ali, B.A. | | 登録日 | 2023-08-29 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | The N1 domain of the peroxisomal AAA-ATPase Pex6 is required for Pex15 binding and proper assembly with Pex1.
J.Biol.Chem., 300, 2023
|
|
8TDO
 
 | |
8TDN
 
 | |
8Q1T
 
 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with IOTA739 | | 分子名称: | 1,10-PHENANTHROLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | 著者 | Cederfelt, D, Lund, B.A, Boronat, P, Hennig, S, Dobritzsch, D, Danielson, U.H. | | 登録日 | 2023-08-01 | | 公開日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
