6S19
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.Ch. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1D
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from 20,000 diffraction patterns | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1G
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from 50,000 diffraction patterns. | | 分子名称: | L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
5NM5
 
 | | Tubulin Darpin room-temperature structure in complex with Colchicine determined by serial millisecond crystallography | | 分子名称: | Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Weinert, T, Olieric, N, James, D, Gashi, D, Nogly, P, Jaeger, K, Steinmetz, M.O, Standfuss, J. | | 登録日 | 2017-04-05 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
4B3M
 
 | | Crystal structure of the 30S ribosome in complex with compound 1 | | 分子名称: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4,6-O-benzylidene-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | 登録日 | 2012-07-25 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
2VTJ
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | 分子名称: | 4-[(6-chloropyrazin-2-yl)amino]benzenesulfonamide, CELL DIVISION PROTEIN KINASE 2, GLYCEROL | | 著者 | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | 登録日 | 2008-05-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
8PZO
 
 | | LpdD | | 分子名称: | Protein LpdD, SODIUM ION | | 著者 | Gahloth, D, Leys, D. | | 登録日 | 2023-07-27 | | 公開日 | 2024-01-17 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The prFMNH 2 -binding chaperone LpdD assists UbiD decarboxylase activation.
J.Biol.Chem., 300, 2024
|
|
2VTI
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | 分子名称: | CELL DIVISION PROTEIN KINASE 2, N-(4-sulfamoylphenyl)-1H-indazole-3-carboxamide | | 著者 | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | 登録日 | 2008-05-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VTR
 
 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design | | 分子名称: | 5-chloro-7-[(1-methylethyl)amino]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | 登録日 | 2008-05-15 | | 公開日 | 2008-08-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
6DMA
 
 | | DHD15_closed | | 分子名称: | DHD15_closed_A, DHD15_closed_B | | 著者 | Bick, M.J, Chen, Z, Baker, D. | | 登録日 | 2018-06-04 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.363 Å) | | 主引用文献 | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
8PO5
 
 | | Lactobacillus plantarum LpdD | | 分子名称: | MANGANESE (II) ION, Protein LpdD | | 著者 | Gahloth, D, Leys, D. | | 登録日 | 2023-07-03 | | 公開日 | 2024-01-17 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The prFMNH 2 -binding chaperone LpdD assists UbiD decarboxylase activation.
J.Biol.Chem., 300, 2024
|
|
8PZH
 
 | | LpdD (H61A) mutant | | 分子名称: | MANGANESE (II) ION, PHOSPHATE ION, Protein LpdD | | 著者 | Gahloth, D, Leys, D. | | 登録日 | 2023-07-27 | | 公開日 | 2024-01-17 | | 最終更新日 | 2025-01-29 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | The prFMNH 2 -binding chaperone LpdD assists UbiD decarboxylase activation.
J.Biol.Chem., 300, 2024
|
|
1E0X
 
 | | XYLANASE 10A FROM SREPTOMYCES LIVIDANS. XYLOBIOSYL-ENZYME INTERMEDIATE AT 1.65 A | | 分子名称: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | 著者 | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | 登録日 | 2000-04-10 | | 公開日 | 2001-04-05 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
4B3R
 
 | | Crystal structure of the 30S ribosome in complex with compound 30 | | 分子名称: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-4,6-O-[(1R)-3-phenylpropylidene]-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | 著者 | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | 登録日 | 2012-07-26 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
6S0Q
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from 50,000 diffraction patterns | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | 著者 | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
5NM4
 
 | | A2A Adenosine receptor room-temperature structure determined by serial femtosecond crystallography | | 分子名称: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | 著者 | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Hennig, M, Standfuss, J. | | 登録日 | 2017-04-05 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
6PPB
 
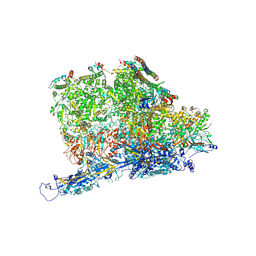 | | Kaposi's sarcoma-associated herpesvirus (KSHV), C5 portal vertex structure | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | 著者 | Gong, D, Dai, X, Jih, J, Liu, Y.T, Bi, G.Q, Sun, R, Zhou, Z.H. | | 登録日 | 2019-07-06 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.
Cell, 178, 2019
|
|
6GF0
 
 | | Lysozyme structure determined from SFX data using a Sheet-on-Sheet chipless chip | | 分子名称: | Lysozyme C | | 著者 | Doak, R.B, Gorel, A, Foucar, L, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrel, D, Owen, R, Barends, T.R.M, Schlichting, I. | | 登録日 | 2018-04-27 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8POK
 
 | | Cryo-EM structure of cell-free synthesized human histamine H2 receptor coupled to heterotrimeric Gs protein in lipid environment | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, HISTAMINE, ... | | 著者 | Schnelle, K, Koeck, Z, Persechino, M, Umbach, S, Schihada, H, Januliene, D, Parey, K, Pockes, S, Kolb, P, Doetsch, V, Moeller, A, Hilger, D, Bernhard, F. | | 登録日 | 2023-07-05 | | 公開日 | 2024-03-06 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structure of cell-free synthesized human histamine 2 receptor/G s complex in nanodisc environment.
Nat Commun, 15, 2024
|
|
4AE8
 
 | | Crystal structure of human THEM4 | | 分子名称: | THIOESTERASE SUPERFAMILY MEMBER 4 | | 著者 | Zhuravleva, E, Gut, H, Hynx, D, Marcellin, D, Bleck, C.K.E, Genoud, C, Cron, P, Keusch, J.J, Dummler, B, Degli Esposti, M, Hemmings, B.A. | | 登録日 | 2012-01-09 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.594 Å) | | 主引用文献 | Acyl Coenzyme a Thioesterase Them5/Acot15 is Involved in Cardiolipin Remodeling and Fatty Liver Development.
Mol.Cell.Biol., 32, 2012
|
|
5AAZ
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | 分子名称: | OPTINEURIN, ZINC ION | | 著者 | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | 登録日 | 2015-07-31 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
4A0Z
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with malonyl-CoA | | 分子名称: | MALONYL-COENZYME A, TRANSCRIPTION FACTOR FAPR | | 著者 | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | 登録日 | 2011-09-13 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
1WOH
 
 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | 分子名称: | agmatinase | | 著者 | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | 登録日 | 2004-08-18 | | 公開日 | 2004-09-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1E0V
 
 | | Xylanase 10A from Sreptomyces lividans. cellobiosyl-enzyme intermediate at 1.7 A | | 分子名称: | ENDO-1,4-BETA-XYLANASE A, beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | 著者 | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | 登録日 | 2000-04-10 | | 公開日 | 2001-04-05 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | 登録日 | 2018-01-30 | | 公開日 | 2018-04-18 | | 最終更新日 | 2025-03-19 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
