7TUZ
 
 | | Cryo-EM structure of 7alpha,25-dihydroxycholesterol-bound EBI2/GPR183 in complex with Gi protein | | 分子名称: | (2S,4aS,4bS,7R,8S,8aS,9R,10aR)-7-[(2R,3R)-7-hydroxy-3,7-dimethyloctan-2-yl]-4a,7,8-trimethyltetradecahydrophenanthrene-2,9-diol, G-protein coupled receptor 183, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Chen, H, Hung, W, Li, X. | | 登録日 | 2022-02-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
7TUY
 
 | | Cryo-EM structure of GSK682753A-bound EBI2/GPR183 | | 分子名称: | 8-[(2E)-3-(4-chlorophenyl)prop-2-enoyl]-3-[(3,4-dichlorophenyl)methyl]-1-oxa-3,8-diazaspiro[4.5]decan-2-one, G-protein coupled receptor 183,Soluble cytochrome b562 fusion, anti-BRIL Fab Heavy chain, ... | | 著者 | Chen, H, Huang, W, Li, X. | | 登録日 | 2022-02-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Structures of oxysterol sensor EBI2/GPR183, a key regulator of the immune response.
Structure, 30, 2022
|
|
3P6Y
 
 | | CF Im25-CF Im68-UGUAA complex | | 分子名称: | 5'-R(*UP*GP*UP*AP*A)-3', Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | 著者 | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | 登録日 | 2010-10-11 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
3P5T
 
 | | CFIm25-CFIm68 complex | | 分子名称: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | 著者 | Li, H, Tong, S, Li, X, Shi, H, Gao, Y, Ge, H, Niu, L, Teng, M. | | 登録日 | 2010-10-11 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of pre-mRNA recognition by the human cleavage factor Im complex
To be Published
|
|
8G94
 
 | | Structure of CD69-bound S1PR1 coupled to heterotrimeric Gi | | 分子名称: | Early activation antigen CD69, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Chen, H, Li, X. | | 登録日 | 2023-02-21 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-04-26 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Transmembrane protein CD69 acts as an S1PR1 agonist.
Elife, 12, 2023
|
|
8G92
 
 | | Structure of inhibitor 16d-bound SPNS2 | | 分子名称: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, Sphingosine-1-phosphate transporter SPNS2 | | 著者 | Chen, H, Li, X. | | 登録日 | 2023-02-21 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
4LA1
 
 | | Crystal structure of SjTGR (thioredoxin glutathione reductase from Schistosoma japonicumi)complex with FAD | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin glutathione reductase | | 著者 | Peng, Y, Wu, Q, Huang, F, Chen, J, Li, X, Zhou, X, Fan, X. | | 登録日 | 2013-06-18 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.348 Å) | | 主引用文献 | Crystal structure of SjTGR complex with FAD
To be Published
|
|
5X0X
 
 | | Complex of Snf2-Nucleosome complex with Snf2 bound to position +6 of the nucleosome | | 分子名称: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | 登録日 | 2017-01-23 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.97 Å) | | 主引用文献 | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
5X0Y
 
 | | Complex of Snf2-Nucleosome complex with Snf2 bound to SHL2 of the nucleosome | | 分子名称: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | 登録日 | 2017-01-23 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.69 Å) | | 主引用文献 | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
3HVN
 
 | | Crystal structure of cytotoxin protein suilysin from Streptococcus suis | | 分子名称: | 1,1,1,3,3,3-hexafluoropropan-2-ol, HEPTANE-1,2,3-TRIOL, Hemolysin | | 著者 | Xu, L, Huang, B, Du, H, Zhang, C.X, Xu, J, Li, X, Rao, Z. | | 登録日 | 2009-06-16 | | 公開日 | 2010-03-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.852 Å) | | 主引用文献 | Crystal structure of cytotoxin protein suilysin from Streptococcus suis.
Protein Cell, 1, 2010
|
|
6LML
 
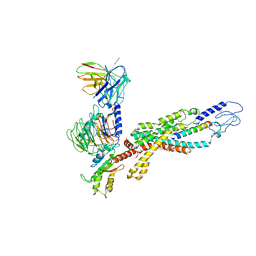 | | Cryo-EM structure of the human glucagon receptor in complex with Gi1 | | 分子名称: | Glucagon, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Qiao, A, Han, S, Li, X, Sun, F, Zhao, Q, Wu, B. | | 登録日 | 2019-12-26 | | 公開日 | 2020-04-01 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of Gsand Girecognition by the human glucagon receptor.
Science, 367, 2020
|
|
4QPG
 
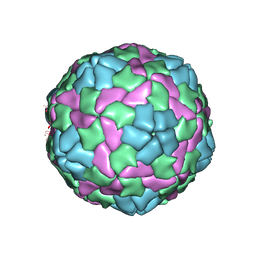 | | Crystal structure of empty hepatitis A virus | | 分子名称: | CHLORIDE ION, Capsid protein VP0, Capsid protein VP1, ... | | 著者 | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | 登録日 | 2014-06-23 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4QPI
 
 | | Crystal structure of hepatitis A virus | | 分子名称: | CHLORIDE ION, Capsid protein VP1, Capsid protein VP2, ... | | 著者 | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | 登録日 | 2014-06-23 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4D1Q
 
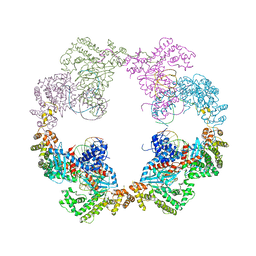 | | Hermes transposase bound to its terminal inverted repeat | | 分子名称: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | 著者 | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | 登録日 | 2014-05-04 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
5TRD
 
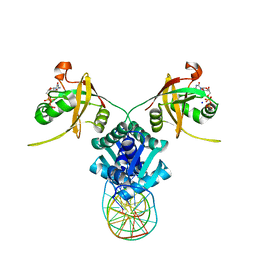 | | Structure of RbkR (Riboflavin Kinase) from Thermoplasma acidophilum determined in complex with CTP and its cognate DNA operator | | 分子名称: | CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*TP*TP*AP*CP*TP*AP*AP*TP*TP*CP*AP*CP*GP*AP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*TP*CP*GP*TP*GP*AP*AP*TP*TP*AP*GP*TP*AP*A)-3'), ... | | 著者 | Vetting, M.W, Rodionova, I.A, Li, X, Osterman, A.L, Rodionov, D.A, Almo, S.C. | | 登録日 | 2016-10-26 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of RbkR (Riboflavin Kinase) from Thermoplasma acidophilum determined in complex with CTP and its cognate DNA operator
To be published
|
|
4EDL
 
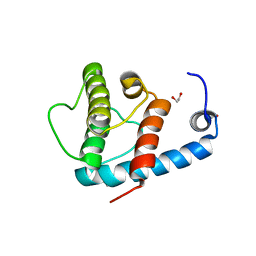 | | Crystal structure of beta-parvin CH2 domain | | 分子名称: | 1,2-ETHANEDIOL, Beta-parvin | | 著者 | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | 登録日 | 2012-03-27 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
4DM4
 
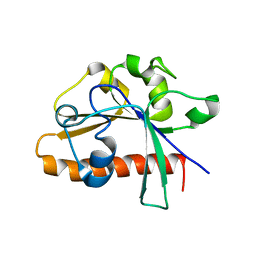 | | The conserved domain of yeast Cdc73 | | 分子名称: | Cell division control protein 73 | | 著者 | Chen, H, Shi, N, Gao, Y, Li, X, Niu, L, Teng, M. | | 登録日 | 2012-02-06 | | 公開日 | 2012-08-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4ED5
 
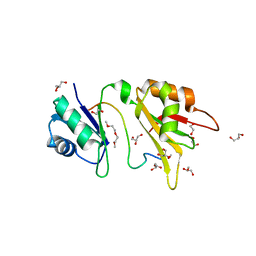 | | Crystal structure of the two N-terminal RRM domains of HuR complexed with RNA | | 分子名称: | 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 5'-R(*A*UP*UP*UP*UP*UP*AP*UP*UP*UP*U)-3', ... | | 著者 | Wang, H, Zeng, F, Liu, Q, Niu, L, Teng, M, Li, X. | | 登録日 | 2012-03-27 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4E7N
 
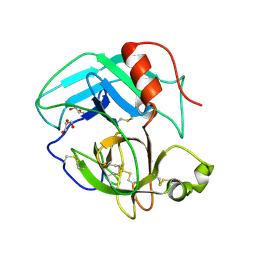 | | Crystal Structure of AhV_TL-I, a Glycosylated Snake-venom Thrombin-like Enzyme from Agkistrodon halys | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Snake-venom Thrombin-like Enzyme | | 著者 | Zeng, F, Li, X, Teng, M, Niu, L. | | 登録日 | 2012-03-18 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure of AhV_TL-I, a Glycosylated Snake-venom Thrombin-like Enzyme from Agkistrodon halys
to be published
|
|
3KXL
 
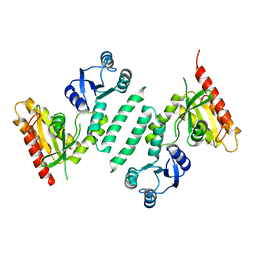 | | crystal structure of SsGBP mutation variant G235S | | 分子名称: | GTP-binding protein (HflX), THIOCYANATE ION | | 著者 | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | 登録日 | 2009-12-03 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
3KXI
 
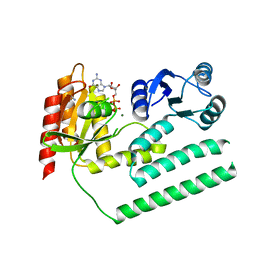 | | crystal structure of SsGBP and GDP complex | | 分子名称: | GTP-binding protein (HflX), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | 登録日 | 2009-12-03 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
3KXK
 
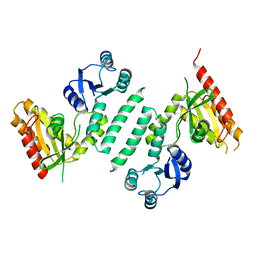 | |
4EDN
 
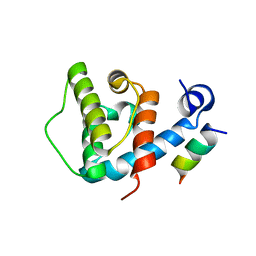 | | Crystal structure of beta-parvin CH2 domain in complex with paxillin LD1 motif | | 分子名称: | Beta-parvin, Paxillin, SULFATE ION | | 著者 | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | 登録日 | 2012-03-27 | | 公開日 | 2012-08-08 | | 最終更新日 | 2013-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
4RM0
 
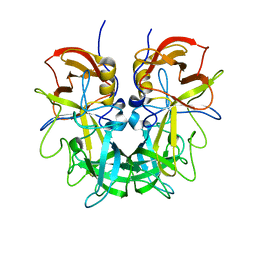 | | Crystal structure of Norovirus OIF P domain in complex with Lewis a trisaccharide | | 分子名称: | Capsid protein, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Liu, W, Chen, Y, Tan, M, Xia, M, Li, X, Jiang, X, Rao, Z. | | 登録日 | 2014-10-18 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface.
Plos Pathog., 11, 2015
|
|
4JGZ
 
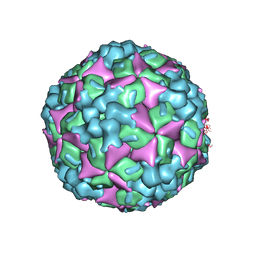 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group I222) | | 分子名称: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | 著者 | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, McAuley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | 登録日 | 2013-03-04 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
