5GVT
 
 | |
7VTC
 
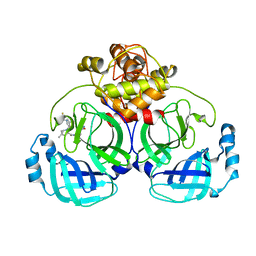 | | Crystal structure of MERS main protease in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2021-10-28 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.53865623 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLQ
 
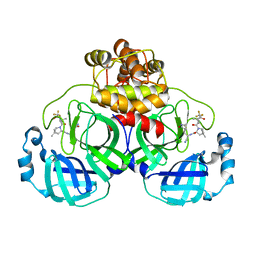 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.939106 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLO
 
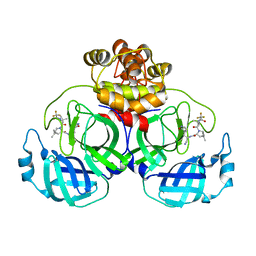 | | Crystal structure of SARS coronavirus main protease in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.0227 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.50251937 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
5ZZ3
 
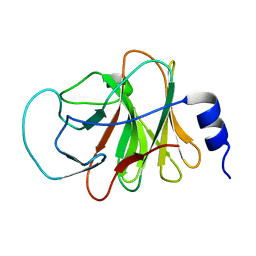 | | Crystal structure of intracellular B30.2 domain of BTN3A3 | | 分子名称: | Butyrophilin, subfamily 3, member A3 isoform b variant | | 著者 | Yang, Y.Y, Li, X, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-05-30 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
8HCI
 
 | |
5IPE
 
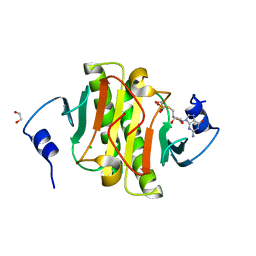 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) nucleoside thiophosphoramidate catalytic product complex | | 分子名称: | 1,2-ETHANEDIOL, 5'-S-phosphono-5'-thioguanosine, CHLORIDE ION, ... | | 著者 | Maize, K.M, Finzel, B.C. | | 登録日 | 2016-03-09 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
5IPB
 
 | |
5IPC
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside thiophosphoramidate substrate complex | | 分子名称: | 1,2-ETHANEDIOL, 5'-S-[(S)-hydroxy{[2-(1H-indol-3-yl)ethyl]amino}phosphoryl]-5'-thioguanosine, CHLORIDE ION, ... | | 著者 | Maize, K.M, Finzel, B.C. | | 登録日 | 2016-03-09 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
5IPD
 
 | |
8HUR
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HI8
 
 | |
8HI7
 
 | |
5JK7
 
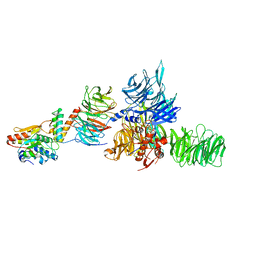 | | The X-ray structure of the DDB1-DCAF1-Vpr-UNG2 complex | | 分子名称: | DNA damage-binding protein 1, Protein VPRBP, Protein Vpr, ... | | 著者 | Calero, G, Ahn, J, Wu, Y. | | 登録日 | 2016-04-26 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.49 Å) | | 主引用文献 | The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7BOK
 
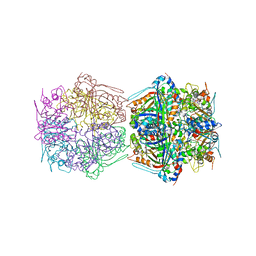 | | Cryo-EM structure of the encapsulated DyP-type peroxidase from Mycobacterium smegmatis | | 分子名称: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BOJ
 
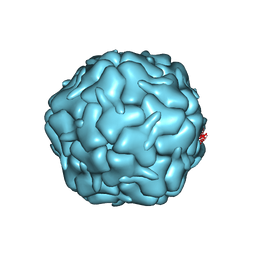 | | Cryo-EM structure of the encapsulin shell from Mycobacterium smegmatis | | 分子名称: | 29 kDa antigen Cfp29 | | 著者 | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | 登録日 | 2020-03-19 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6JIQ
 
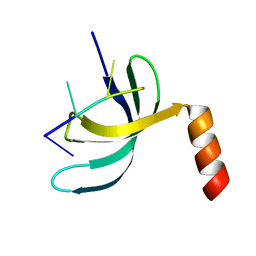 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | 分子名称: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | 著者 | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | 登録日 | 2019-02-22 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6JIP
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | 分子名称: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), PENTAETHYLENE GLYCOL, SP_0782 | | 著者 | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | 登録日 | 2019-02-22 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.659 Å) | | 主引用文献 | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
8W4U
 
 | | human KCNQ2-CaM in complex with PIP2 and HN37 | | 分子名称: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | 著者 | Ma, D, Li, X, Guo, J. | | 登録日 | 2023-08-25 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
5ZXK
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP | | 分子名称: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1 | | 著者 | Yang, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2018-05-21 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
5ZKM
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA TCTTCC | | 分子名称: | DNA (5'-D(P*TP*CP*TP*TP*CP*C)-3'), SP_0782 | | 著者 | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | 登録日 | 2018-03-24 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
5ZKL
 
 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | 分子名称: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | 著者 | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | 登録日 | 2018-03-24 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
7CMU
 
 | | Dopamine Receptor D3R-Gi-Pramipexole complex | | 分子名称: | (6S)-N6-propyl-4,5,6,7-tetrahydro-1,3-benzothiazole-2,6-diamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | 登録日 | 2020-07-29 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
7CMV
 
 | | Dopamine Receptor D3R-Gi-PD128907 complex | | 分子名称: | (4aR,10bR)-4-propyl-3,4a,5,10b-tetrahydro-2H-chromeno[4,3-b][1,4]oxazin-9-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Xu, P, Huang, S, Mao, C, Krumm, B, Zhou, X, Tan, Y, Huang, X.-P, Liu, Y, Shen, D.-D, Jiang, Y, Yu, X, Jiang, H, Melcher, K, Roth, B, Cheng, X, Zhang, Y, Xu, H. | | 登録日 | 2020-07-29 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structures of the human dopamine D3 receptor-G i complexes.
Mol.Cell, 81, 2021
|
|
