8SUR
 
 | | TMEM16F bound with Niclosamide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-chloro-4-nitrophenyl)-2-hydroxybenzamide, ... | | 著者 | Feng, S, Cheng, Y. | | 登録日 | 2023-05-13 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8TAG
 
 | | TMEM16F, with Calcium and PIP2, no inhibitor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, CALCIUM ION | | 著者 | Feng, S, Cheng, Y. | | 登録日 | 2023-06-27 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
4OI3
 
 | | Crystal structure analysis of SCO4226 from Streptomyces coelicolor A3(2) | | 分子名称: | Nickel responsive protein | | 著者 | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | 登録日 | 2014-01-18 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
8SUN
 
 | | TMEM16F 1PBC | | 分子名称: | 1-Hydroxy-3-(trifluoromethyl)pyrido[1,2-a]benzimidazole-4-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, ... | | 著者 | Wu, H, Feng, S, Cheng, Y. | | 登録日 | 2023-05-12 | | 公開日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8TAI
 
 | | TMEM16F, with Calcium and PIP2, no inhibitor, Cl2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, CALCIUM ION | | 著者 | Wu, H, Feng, S, Cheng, Y. | | 登録日 | 2023-06-27 | | 公開日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8TAL
 
 | | TMEM16F, with Calcium and PIP2, no inhibitor, Cl1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, CALCIUM ION | | 著者 | Wu, H, Feng, S, Cheng, Y. | | 登録日 | 2023-06-27 | | 公開日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
4ERG
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, FE (III) ION | | 著者 | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | 登録日 | 2012-04-20 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.789 Å) | | 主引用文献 | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4ERI
 
 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | 著者 | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | 登録日 | 2012-04-20 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.0006 Å) | | 主引用文献 | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
4EPK
 
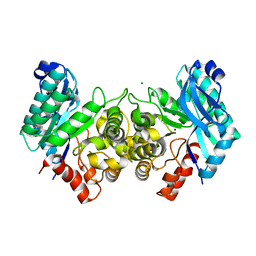 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | 著者 | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | 登録日 | 2012-04-17 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6009 Å) | | 主引用文献 | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
3Q2C
 
 | | Binding properties to HLA class I molecules and the structure of the leukocyte Ig-like receptor A3 (LILRA3/ILT6/LIR4/CD85e) | | 分子名称: | Leukocyte immunoglobulin-like receptor subfamily A member 3 | | 著者 | Ryu, M, Chen, Y, Qi, J.X, Liu, J, Shi, Y, Cheng, H, Gao, G.F. | | 登録日 | 2010-12-20 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence
Plos One, 6, 2011
|
|
4ERA
 
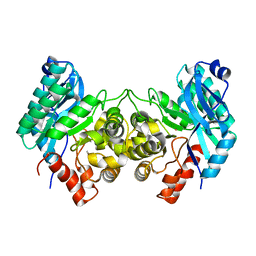 | | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase | | 分子名称: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | 著者 | Huo, L, Fielding, A.J, Chen, Y, Li, T, Iwaki, H, Hosler, J.P, Chen, L, Hasegawa, Y, Que Jr, L, Liu, A. | | 登録日 | 2012-04-19 | | 公開日 | 2012-08-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.398 Å) | | 主引用文献 | Evidence for a Dual Role of an Active Site Histidine in alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase
Biochemistry, 51, 2012
|
|
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | 分子名称: | ATP-dependent DNA helicase II, 80 kDa subunit | | 著者 | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | 登録日 | 2003-12-15 | | 公開日 | 2003-12-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
8HBM
 
 | |
4FKZ
 
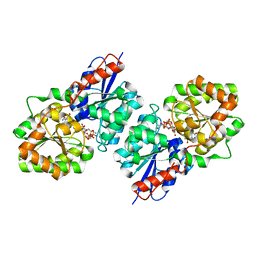 | | Crystal structure of Bacillus subtilis UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP | | 分子名称: | UDP-N-acetylglucosamine 2-epimerase, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Yang, C.S, Chen, S.C, Kuan, S.M, Chen, Y.R, Liu, Y.H, Chen, Y. | | 登録日 | 2012-06-14 | | 公開日 | 2013-05-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Crystal structure of Bacillus subtilis UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP
To be Published
|
|
1IG6
 
 | | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES | | 分子名称: | MODULATOR RECOGNITION FACTOR 2 | | 著者 | Lin, D, Tsui, V, Case, D, Yuan, Y.C, Chen, Y. | | 登録日 | 2001-04-17 | | 公開日 | 2001-04-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES
To be Published
|
|
5BND
 
 | |
8DDI
 
 | |
8DDM
 
 | |
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
6K5E
 
 | |
7RN0
 
 | |
7RN1
 
 | |
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
