3DEI
 
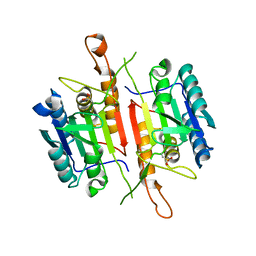 | | Crystal Structures of Caspase-3 with Bound Isoquinoline-1,3,4-trione Derivative Inhibitors | | 分子名称: | (1S)-2-oxo-1-phenyl-2-[(1,3,4-trioxo-1,2,3,4-tetrahydroisoquinolin-5-yl)amino]ethyl acetate, Caspase-3 | | 著者 | Wu, J, Du, J, Li, J, Ding, J. | | 登録日 | 2008-06-10 | | 公開日 | 2008-09-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Isoquinoline-1,3,4-trione Derivatives Inactivate Caspase-3 by Generation of Reactive Oxygen Species
J.Biol.Chem., 283, 2008
|
|
2EW7
 
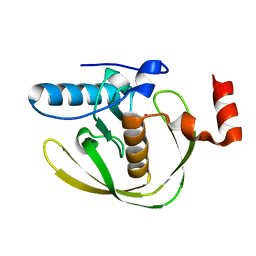 | | Crystal Structure of Helicobacter Pylori peptide deformylase | | 分子名称: | COBALT (II) ION, peptide deformylase | | 著者 | Cai, J. | | 登録日 | 2005-11-02 | | 公開日 | 2006-10-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2EW6
 
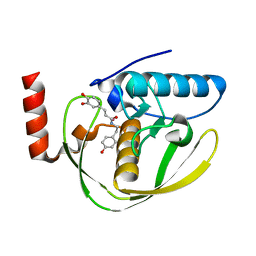 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | 分子名称: | (2E)-3-(3,4-DIHYDROXYPHENYL)-N-[2-(4-HYDROXYPHENYL)ETHYL]ACRYLAMIDE, COBALT (II) ION, peptide deformylase | | 著者 | Cai, J. | | 登録日 | 2005-11-02 | | 公開日 | 2006-10-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2EW5
 
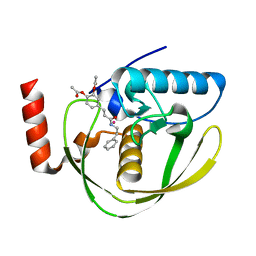 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | 分子名称: | 4-{(1E)-3-OXO-3-[(2-PHENYLETHYL)AMINO]PROP-1-EN-1-YL}-1,2-PHENYLENE DIACETATE, COBALT (II) ION, peptide deformylase | | 著者 | Cai, J. | | 登録日 | 2005-11-02 | | 公開日 | 2006-10-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2F7E
 
 | | PKA complexed with (S)-2-(1H-Indol-3-yl)-1-(5-isoquinolin-6-yl-pyridin-3-yloxymethyl-etylamine | | 分子名称: | (1S)-2-(1H-INDOL-3-YL)-1-{[(5-ISOQUINOLIN-6-YLPYRIDIN-3-YL)OXY]METHYL}ETHYLAMINE, PKI inhibitory peptide, cAMP-dependent protein kinase, ... | | 著者 | Stoll, V.S. | | 登録日 | 2005-11-30 | | 公開日 | 2006-06-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Synthesis and structure-activity relationship of 3,4'-bispyridinylethylenes: discovery of a potent 3-isoquinolinylpyridine inhibitor of protein kinase B (PKB/Akt) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1S9E
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | 分子名称: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | 著者 | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | 登録日 | 2004-02-04 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9G
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | 分子名称: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | 著者 | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | 登録日 | 2004-02-04 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
6VWV
 
 | |
6VWT
 
 | |
1SUQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R185545 | | 分子名称: | (6-[4-(AMINOMETHYL)-2,6-DIMETHYLPHENOXY]-2-{[4-(AMINOMETHYL)PHENYL]AMINO}-5-BROMOPYRIMIDIN-4-YL)METHANOL, MAGNESIUM ION, REVERSE TRANSCRIPTASE | | 著者 | Das, K, Arnold, E. | | 登録日 | 2004-03-26 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1S6Q
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | 分子名称: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | 著者 | Das, K, Arnold, E. | | 登録日 | 2004-01-26 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1J5O
 
 | | CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER | | 分子名称: | 5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3', ANTIBODY (HEAVY CHAIN), ... | | 著者 | Sarafianos, S.G, Das, K, Arnold, E. | | 登録日 | 2002-05-24 | | 公開日 | 2002-06-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2IAJ
 
 | |
2IC3
 
 | |
1SV5
 
 | | CRYSTAL STRUCTURE OF K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R165335 | | 分子名称: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse Transcriptase | | 著者 | Das, K, Arnold, E. | | 登録日 | 2004-03-27 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
3B8W
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221P | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-11-02 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8T
 
 | | Crystal structure of Escherichia coli alaine racemase mutant P219A | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-11-02 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8V
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221K | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-11-02 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
3B8U
 
 | | Crystal structure of Escherichia coli alaine racemase mutant E221A | | 分子名称: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | 登録日 | 2007-11-02 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
1S6P
 
 | |
4EKF
 
 | |
3BLI
 
 | |
2I5J
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with DHBNH, an RNASE H inhibitor | | 分子名称: | (E)-3,4-DIHYDROXY-N'-[(2-METHOXYNAPHTHALEN-1-YL)METHYLENE]BENZOHYDRAZIDE, MAGNESIUM ION, Reverse transcriptase/ribonuclease H P51 subunit, ... | | 著者 | Himmel, D.M, Sarafianos, S.G, Knight, J.L, Levy, R.M, Arnold, E. | | 登録日 | 2006-08-24 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site.
Acs Chem.Biol., 1, 2006
|
|
2ZE2
 
 | | Crystal structure of L100I/K103N mutant HIV-1 reverse transcriptase (RT) in complex with TMC278 (rilpivirine), a non-nucleoside RT inhibitor | | 分子名称: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | 著者 | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | 登録日 | 2007-12-05 | | 公開日 | 2008-02-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8GT6
 
 | | human STING With agonist HB3089 | | 分子名称: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | 著者 | Wang, Z, Yu, X. | | 登録日 | 2022-09-07 | | 公開日 | 2022-12-28 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
