6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | 分子名称: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2018-05-31 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
6R48
 
 | |
6R46
 
 | |
4F7G
 
 | |
5X26
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with SKLB(3) | | 分子名称: | CHLORIDE ION, Epidermal growth factor receptor, N2-[4-(4-methylpiperazin-1-yl)phenyl]-N8-phenyl-9-propan-2-yl-purine-2,8-diamine | | 著者 | Yun, C.H. | | 登録日 | 2017-01-31 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.951 Å) | | 主引用文献 | Structural insights into drug development strategy targeting EGFR T790M/C797S.
Oncotarget, 9, 2018
|
|
5X2K
 
 | |
4FHA
 
 | | Structure of Dihydrodipicolinate Synthase from Streptococcus pneumoniae,bound to pyruvate and lysine | | 分子名称: | Dihydrodipicolinate synthase, LYSINE, SODIUM ION | | 著者 | Perugini, M.A, Dogovski, C, Parker, M.W, Gorman, M.A. | | 登録日 | 2012-06-06 | | 公開日 | 2013-09-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structure, Function, Stability and Knockout Phenotype of Dihydrodipicolinate Synthase from Streptococcus pneumoniae
To be Published
|
|
7YV4
 
 | | Crystal structure of human UCHL3 in complex with Farrerol | | 分子名称: | (2~{S})-2-(4-hydroxyphenyl)-6,8-dimethyl-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | 著者 | Mao, Z.Y, Xu, X.J, Zhang, W.T. | | 登録日 | 2022-08-18 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Farrerol directly activates the deubiqutinase UCHL3 to promote DNA repair and reprogramming when mediated by somatic cell nuclear transfer.
Nat Commun, 14, 2023
|
|
6DXL
 
 | | Linked amidobenzimidazole STING agonist | | 分子名称: | 1,1'-(butane-1,4-diyl)bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1H-benzimidazole-5-carboxamide}, CALCIUM ION, Stimulator of interferon protein | | 著者 | Concha, N.O. | | 登録日 | 2018-06-29 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
6DXG
 
 | | amidobenzimidazole (ABZI) STING agonists | | 分子名称: | 2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-1-[(2R)-2-hydroxy-2-phenylethyl]-1H-benzimidazole-5-carboxamide, CALCIUM ION, Stimulator of interferon protein | | 著者 | Concha, N.O. | | 登録日 | 2018-06-28 | | 公開日 | 2018-11-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.905 Å) | | 主引用文献 | Design of amidobenzimidazole STING receptor agonists with systemic activity.
Nature, 564, 2018
|
|
4XS2
 
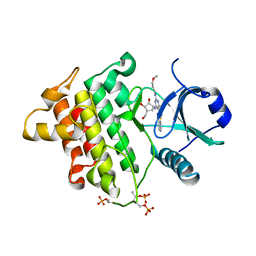 | | Irak4-inhibitor co-structure | | 分子名称: | (1R,2S,3R,5R)-3-({5-(1,3-benzothiazol-2-yl)-6-chloro-2-[(3-methoxypropyl)amino]pyrimidin-4-yl}amino)-5-(hydroxymethyl)cyclopentane-1,2-diol, Interleukin-1 receptor-associated kinase 4 | | 著者 | Fischmann, T.O. | | 登録日 | 2015-01-21 | | 公開日 | 2015-05-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Discovery and hit-to-lead optimization of 2,6-diaminopyrimidine inhibitors of interleukin-1 receptor-associated kinase 4.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3BVG
 
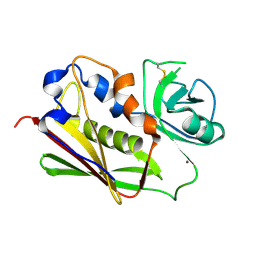 | |
3C8F
 
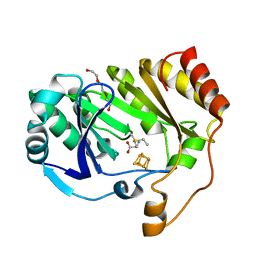 | |
3BYT
 
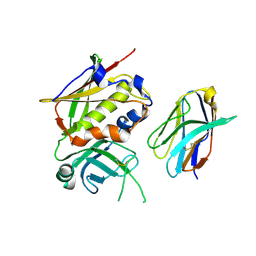 | |
3BVZ
 
 | |
6X7V
 
 | |
6X7U
 
 | |
8W9W
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide/phosphoethanolamine | | 分子名称: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1, ~{N}-[(~{Z},2~{S},3~{R})-1,3-bis(oxidanyl)heptadec-4-en-2-yl]dodecanamide | | 著者 | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | 登録日 | 2023-09-06 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 2024
|
|
8W9Y
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein | | 分子名称: | Sphingomyelin synthase-related protein 1 | | 著者 | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | 登録日 | 2023-09-06 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 2024
|
|
6YMQ
 
 | | TREM2 extracellular domain (19-131) in complex with single-chain variable 4 (scFv-4) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Single-chain variable 4, ... | | 著者 | Szykowska, A, Preger, C, Scacioc, A, Mukhopadhyay, S.M.M, McKinley, G, Graslund, S, Wigren, E, Persson, H, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Di Daniel, E, Davis, J.B, Burgess-Brown, N, Bullock, A. | | 登録日 | 2020-04-09 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
6Y6C
 
 | | TREM2 extracellular domain (19-174) in complex with single-chain variable fragment (scFv-4) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Single chain variable, Triggering receptor expressed on myeloid cells 2 | | 著者 | Szykowska, A, Preger, C, Williams, E, Mukhopadhyay, S.M.M, McKinley, G, Gruslund, S, Wigren, E, Persson, H, Arrowsmith, C.H, Edwards, A, von Delft, F, Bountra, C, Davis, J.B, Di Daniel, E, Burgess-Brown, N, Bullock, A. | | 登録日 | 2020-02-26 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Selection and structural characterization of anti-TREM2 scFvs that reduce levels of shed ectodomain.
Structure, 29, 2021
|
|
6LHU
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | 分子名称: | Fanconi anemia complementation group A | | 著者 | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | 登録日 | 2019-12-10 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHW
 
 | | Structure of N-terminal and C-terminal domains of FANCA | | 分子名称: | Fanconi anemia complementation group A | | 著者 | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | 登録日 | 2019-12-10 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.84 Å) | | 主引用文献 | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHS
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | 分子名称: | Fanconi anemia complementation group A | | 著者 | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | 登録日 | 2019-12-10 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
5XBL
 
 | | Structure of nuclease in complex with associated protein | | 分子名称: | Associated protein, CRISPR-associated endonuclease Cas9/Csn1, RNA (98-MER) | | 著者 | Dong, D, Guo, M, Wang, S, Zhu, Y, Huang, Z. | | 登録日 | 2017-03-20 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.052 Å) | | 主引用文献 | Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein
Nature, 546, 2017
|
|
