7DVM
 
 | |
8YRH
 
 | | Complex of SARS-CoV-2 main protease and Rosmarinic acid | | 分子名称: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, 3C-like proteinase nsp5 | | 著者 | Wang, Q.S, Li, Q.H. | | 登録日 | 2024-03-21 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.841 Å) | | 主引用文献 | Structural basis of rosmarinic acid inhibitory mechanism on SARS-CoV-2 main protease.
Biochem.Biophys.Res.Commun., 724, 2024
|
|
8ZC6
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (6.85 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC1
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | 分子名称: | Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, Spike protein S1 | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-15 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
4FI9
 
 | | Structure of human SUN-KASH complex | | 分子名称: | Nesprin-2, SUN domain-containing protein 2 | | 著者 | Wang, W.J, Shi, Z.B. | | 登録日 | 2012-06-08 | | 公開日 | 2012-07-18 | | 最終更新日 | 2013-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural insights into SUN-KASH complexes across the nuclear envelope.
Cell Res., 22, 2012
|
|
6W51
 
 | |
8Z9A
 
 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum bound with geranyl acetate | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | 著者 | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | 登録日 | 2024-04-23 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
8Z9Z
 
 | | Cryo-EM structure of the insect olfactory receptor OR5-Orco heterocomplex from Acyrthosiphon pisum | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Odorant receptor, ApisOR5, ... | | 著者 | Wang, Y.D, Qiu, L, Guan, Z.Y, Wang, Q, Wang, G.R, Yin, P. | | 登録日 | 2024-04-24 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex.
Science, 384, 2024
|
|
3UNP
 
 | | Structure of human SUN2 SUN domain | | 分子名称: | ACETYL GROUP, SUN domain-containing protein 2 | | 著者 | Zhou, Z.C, Greene, M.I. | | 登録日 | 2011-11-16 | | 公開日 | 2011-12-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope
J.Biol.Chem., 287, 2012
|
|
8Y65
 
 | | Cryo-EM structure of human urate transporter GLUT9 bound to substrate urate | | 分子名称: | Solute carrier family 2, facilitated glucose transporter member 9, URIC ACID | | 著者 | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | 登録日 | 2024-02-01 | | 公開日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8Y66
 
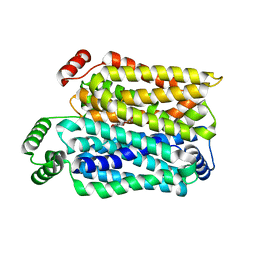 | | Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin | | 分子名称: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Solute carrier family 2, facilitated glucose transporter member 9 | | 著者 | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | 登録日 | 2024-02-01 | | 公開日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
8XS3
 
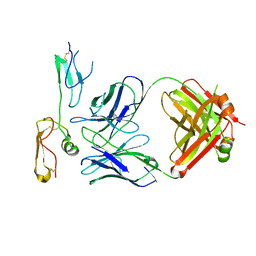 | | Structure of MPXV B6 and D68 fab complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D68_heavy chain, ... | | 著者 | wu, L.L, Sun, J.Q. | | 登録日 | 2024-01-08 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Two noncompeting human neutralizing antibodies targeting MPXV B6 show protective effects against orthopoxvirus infections.
Nat Commun, 15, 2024
|
|
1MFG
 
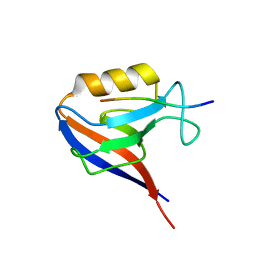 | |
1MFL
 
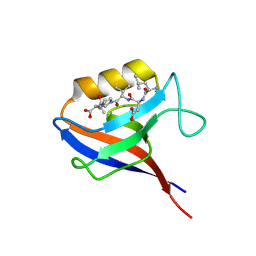 | |
4TW0
 
 | | Crystal Structure of SCARB2 in Acidic Condition (pH4.8) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Dang, M.H, Wang, X.X, Rao, Z.H. | | 登録日 | 2014-06-29 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.648 Å) | | 主引用文献 | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
4TW2
 
 | | Crystal Structure of SCARB2 in Neural Condition (pH7.5) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Scavenger receptor class B member 2, ... | | 著者 | Dang, M.H, Wang, X.X, Rao, Z.H. | | 登録日 | 2014-06-29 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.889 Å) | | 主引用文献 | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
7BW4
 
 | | Structure of the RNA-dependent RNA polymerase from SARS-CoV-2 | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | 著者 | Peng, Q, Peng, R, Shi, Y. | | 登録日 | 2020-04-13 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2.
Cell Rep, 31, 2020
|
|
7RDX
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | 著者 | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | 登録日 | 2021-07-12 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE1
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (composite) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | 著者 | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | 登録日 | 2021-07-12 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.91 Å) | | 主引用文献 | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDZ
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - apo class | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | 著者 | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | 登録日 | 2021-07-12 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE3
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | 著者 | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | 登録日 | 2021-07-12 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE2
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | 著者 | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | 登録日 | 2021-07-12 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | 著者 | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | 登録日 | 2021-07-12 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | 著者 | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | 登録日 | 2021-07-12 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5MEY
 
 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | 登録日 | 2016-11-16 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
