5JP9
 
 | |
6XDG
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | 分子名称: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | 著者 | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | 登録日 | 2020-06-10 | | 公開日 | 2020-06-24 | | 最終更新日 | 2021-01-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
8IH4
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN2A2 Mutant | | 分子名称: | Butyrophilin subfamily 2 member A2 | | 著者 | Yang, Y.Y, Yi, S.M, Zhang, M.T, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2023-02-22 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IGT
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN2A1 | | 分子名称: | Butyrophilin subfamily 2 member A1, ZINC ION | | 著者 | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | 登録日 | 2023-02-21 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
6W9H
 
 | |
6W9I
 
 | |
1QLY
 
 | | NMR Study of the SH3 Domain From Bruton's Tyrosine Kinase, 20 Structures | | 分子名称: | TYROSINE-PROTEIN KINASE BTK | | 著者 | Tzeng, S.R, Lou, Y.C, Pai, M.T, Chen, C, Chen, S.H, Cheng, J.Y. | | 登録日 | 1999-09-20 | | 公開日 | 1999-12-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Human Btk SH3 Domain Complexed with a Proline-Rich Peptide from P120Cbl
J.Biomol.NMR, 16, 2000
|
|
6NSL
 
 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH Compound-6c AKA 6-((1-(4-CYANOPHENY L)-2-OXO-1,2-DIHYDRO-3-PYRIDINYL)AMINO)-N-CYCLOPROPYL-8-(M ETHYLAMINO)IMIDAZO[1,2-B]PYRIDAZINE-3-CARBOXAMIDE | | 分子名称: | 6-{[1-(4-cyanophenyl)-2-oxo-1,2-dihydropyridin-3-yl]amino}-N-cyclopropyl-8-(methylamino)imidazo[1,2-b]pyridazine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | 著者 | Muckelbauer, J.M, Khan, J.A. | | 登録日 | 2019-01-25 | | 公開日 | 2020-01-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Identification of Imidazo[1,2-b]pyridazine Derivatives as Potent, Selective, and Orally Active Tyk2 JH2 Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
2BO3
 
 | | Crystal Structure of HP0242, a Hypothetical Protein from Helicobacter pylori | | 分子名称: | HYPOTHETICAL PROTEIN HP0242 | | 著者 | Sun, Y.-J, Tsai, J.-Y, Chen, B.-T. | | 登録日 | 2005-04-07 | | 公開日 | 2006-06-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal Structure of Hp0242, a Hypothetical Protein from Helicobacter Pylori with a Novel Fold
Proteins: Struct., Funct., Bioinf., 62, 2006
|
|
5MVS
 
 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | 分子名称: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | 著者 | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | 登録日 | 2017-01-17 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW4
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD7 [N-(3-(((R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl)(methyl)amino)propyl)-2-(3-(2-chloro-3-(2-methylpyridin-3-yl)benzo[b]thiophen-5-yl)ureido)acetamide] | | 分子名称: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~2~-{[2-chloro-3-(2-methylpyridin-3-yl)-1-benzothiophen-5-yl]carbamoyl}-N-(3-{methyl[(3R)-1-(5H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]amino}propyl)glycinamide, ... | | 著者 | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | 登録日 | 2017-01-18 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW3
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | 分子名称: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | 著者 | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | 登録日 | 2017-01-18 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
4XKL
 
 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | 分子名称: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | 著者 | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | 登録日 | 2015-01-12 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
6CDE
 
 | | Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | 著者 | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | 登録日 | 2018-02-08 | | 公開日 | 2018-05-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDO
 
 | | Structure of vaccine-elicited HIV-1 neutralizing antibody vFP16.02 in complex with HIV-1 fusion peptide residue 512-519 | | 分子名称: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP16.02 Fab heavy chain, ... | | 著者 | Xu, K, Liu, K, Kwong, P.D. | | 登録日 | 2018-02-08 | | 公開日 | 2018-05-16 | | 最終更新日 | 2018-06-20 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
6CDP
 
 | | Vaccine-elicited HIV-1 neutralizing antibody vFP20.01 in complex with HIV-1 fusion peptide residue 512-519 | | 分子名称: | HIV-1 fusion peptide 512-519, SULFATE ION, vFP20.01 Fab heavy chain, ... | | 著者 | Xu, K, Liu, K, Kwong, P.D. | | 登録日 | 2018-02-08 | | 公開日 | 2018-05-16 | | 最終更新日 | 2018-06-20 | | 実験手法 | X-RAY DIFFRACTION (2.456 Å) | | 主引用文献 | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
3BY7
 
 | |
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | 分子名称: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-04-13 | | 公開日 | 2004-08-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
6W4Q
 
 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | 分子名称: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | 著者 | Greenfield, J, Herzberg, O. | | 登録日 | 2020-03-11 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
6VBK
 
 | | Crystal structure of N-terminal domain of Mycobacterium tuberculosis complex Lon protease | | 分子名称: | GLYCEROL, Lon211 | | 著者 | Bi, F.K, Chen, C, Chen, X.Y, Guo, C.Y, Lin, D.H. | | 登録日 | 2019-12-19 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
4WD5
 
 | |
4WK8
 
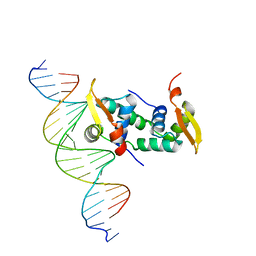 | | FOXP3 forms a domain-swapped dimer to bridge DNA | | 分子名称: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*G)-3'), Forkhead box protein P3 | | 著者 | Chen, Y, Chen, L. | | 登録日 | 2014-10-01 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.4006 Å) | | 主引用文献 | DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions.
Nucleic Acids Res., 43, 2015
|
|
3BOS
 
 | |
3DEE
 
 | |
3CM1
 
 | |
