7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDG
 
 | | Crystal structure of human SARS2 catalytic domain with a disease related mutation | | 分子名称: | Serine--tRNA ligase, mitochondrial | | 著者 | Wu, S, Li, P, Zhou, X.L, Fang, P. | | 登録日 | 2022-07-04 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Selective degradation of tRNASer(AGY) is the primary driver for mitochondrial seryl-tRNA synthetase-related disease.
Nucleic Acids Res., 50, 2022
|
|
7YDF
 
 | | Crystal structure of human SARS2 catalytic domain | | 分子名称: | Serine--tRNA ligase, mitochondrial | | 著者 | Wu, S, Li, P, Zhou, X.L, Fang, P. | | 登録日 | 2022-07-04 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Selective degradation of tRNASer(AGY) is the primary driver for mitochondrial seryl-tRNA synthetase-related disease.
Nucleic Acids Res., 50, 2022
|
|
3C2T
 
 | | Evolution of chlorella virus dUTPase | | 分子名称: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | 著者 | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | 登録日 | 2008-01-25 | | 公開日 | 2009-02-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7FI4
 
 | | Structure of AcrIF13 | | 分子名称: | AcrIF13 | | 著者 | Feng, Y, Gao, T. | | 登録日 | 2021-07-30 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | Mechanistic insights into the inhibition of the CRISPR-Cas surveillance complex by anti-CRISPR protein AcrIF13.
J.Biol.Chem., 298, 2022
|
|
3CA9
 
 | | Evolution of chlorella virus dUTPase | | 分子名称: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | 著者 | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | 登録日 | 2008-02-19 | | 公開日 | 2009-03-03 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7EA8
 
 | |
6IVE
 
 | |
7EA5
 
 | |
7E5R
 
 | | SARS-CoV-2 S trimer with three-antibody cocktail complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | 著者 | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | 登録日 | 2021-02-20 | | 公開日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
7E5S
 
 | | SARS-CoV-2 S trimer with four-antibody cocktail complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | 著者 | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | 登録日 | 2021-02-20 | | 公開日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
6IYW
 
 | | Crystal sturucture of L,D-transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with Imipenem adduct | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, GLYCEROL, L,D-transpeptidase 2 | | 著者 | Li, D.F, Zhao, F, Wang, D.C. | | 登録日 | 2018-12-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The 1-beta-methyl group confers a lower affinity of l,d-transpeptidase LdtMt2 for ertapenem than for imipenem.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6IYY
 
 | |
7EXD
 
 | | Lasmiditan-bound serotonin 1F (5-HT1F) receptor-Gi protein complex | | 分子名称: | 2,4,6-tris(fluoranyl)-N-[6-(1-methylpiperidin-4-yl)carbonylpyridin-2-yl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Huang, S, Xu, P, Jiang, Y, Xu, H.E. | | 登録日 | 2021-05-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for recognition of anti-migraine drug lasmiditan by the serotonin receptor 5-HT 1F -G protein complex.
Cell Res., 31, 2021
|
|
6HU9
 
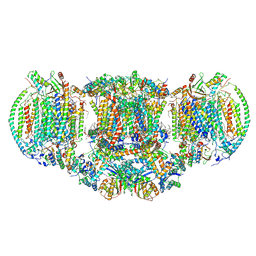 | | III2-IV2 mitochondrial respiratory supercomplex from S. cerevisiae | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CALCIUM ION, ... | | 著者 | Hartley, A.M, Pinotsis, N, Marechal, A. | | 登録日 | 2018-10-05 | | 公開日 | 2018-12-26 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1.
Nat. Struct. Mol. Biol., 26, 2019
|
|
7W6A
 
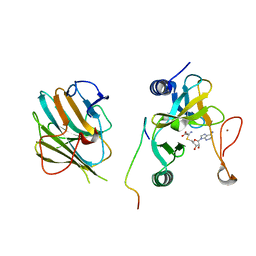 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | 分子名称: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
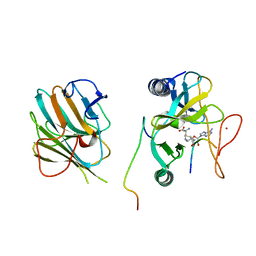 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | 分子名称: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | 著者 | Zhao, L, Li, Y, Chen, Y. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
6JW4
 
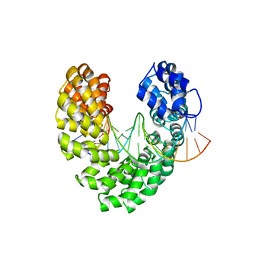 | | Degenerate RVD RG forms a distinct loop conformation | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JW5
 
 | | RVD Q* recognizes 5hmC through water-mediated H bonds | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JW0
 
 | | Universal RVD R* accommodates cytosine via water-mediated interactions | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*CP*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | 分子名称: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | 著者 | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | 登録日 | 2008-11-27 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
6H8P
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Histone H1.4(18-32)K26me3 peptide (15-mer) | | 分子名称: | CHLORIDE ION, GLYCEROL, Histone H1.4, ... | | 著者 | Chowdhury, R, Walport, L.J, Schofield, C.J. | | 登録日 | 2018-08-03 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.983 Å) | | 主引用文献 | Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26.
FEBS Lett., 592, 2018
|
|
6JW3
 
 | | Degenerate RVD RG forms a distinct loop conformation | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
