7Y76
 
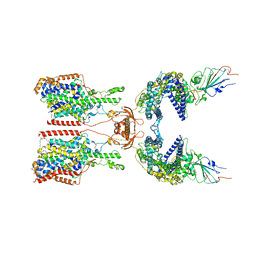 | | SIT1-ACE2-BA.5 RBD | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Shen, Y.P, Li, Y.N, Zhang, Y.Y, Yan, R.H. | | 登録日 | 2022-06-21 | | 公開日 | 2023-01-04 | | 最終更新日 | 2023-02-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of ACE2-SIT1 recognized by Omicron variants of SARS-CoV-2.
Cell Discov, 8, 2022
|
|
8JHF
 
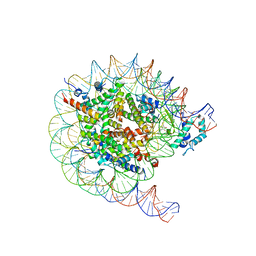 | | Native SUV420H1 bound to 167-bp nucleosome | | 分子名称: | DNA (160-MER), Histone H2A.Z, Histone H2B type 1-K, ... | | 著者 | Lin, F, Li, W. | | 登録日 | 2023-05-23 | | 公開日 | 2023-11-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.68 Å) | | 主引用文献 | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
8JHG
 
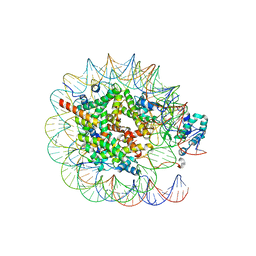 | | Native SUV420H1 bound to 167-bp nucleosome | | 分子名称: | DNA (160-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | 著者 | Lin, F, Li, W. | | 登録日 | 2023-05-23 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
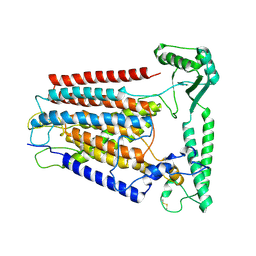
8K0B
 
 | |
7VKK
 
 | |
7VKR
 
 | |
7VKQ
 
 | |
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Wei, H, Guo, M, Wang, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Guo, M, Wei, H, Wang, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.354 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Guo, M, Wang, H, Wei, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.522 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
7WDM
 
 | | Fungal immunomodulatory protein FIP-gmi | | 分子名称: | Immunomodulatory protein | | 著者 | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | 登録日 | 2021-12-22 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.123 Å) | | 主引用文献 | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties
To Be Published
|
|
8IBV
 
 | | Cryo-EM structure of the motilin-bound motilin receptor-Gq protein complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | 著者 | Jiang, Y, Xu, H.E, You, C, Xu, Y. | | 登録日 | 2023-02-10 | | 公開日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
8IBU
 
 | | Cryo-EM structure of the erythromycin-bound motilin receptor-Gq protein complex | | 分子名称: | ERYTHROMYCIN A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | You, C, Jiang, Y, Xu, H.E, Xu, Y. | | 登録日 | 2023-02-10 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | Structural basis for motilin and erythromycin recognition by motilin receptor.
Sci Adv, 9, 2023
|
|
7VR1
 
 | |
7XP1
 
 | | Crystal structure of PmiR from Pseudomonas aeruginosa | | 分子名称: | ALPHA-METHYLISOCITRIC ACID, GLYCEROL, Probable transcriptional regulator, ... | | 著者 | Zhang, Y.X, Liang, H.H, Gan, J.H. | | 登録日 | 2022-05-02 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | PmiR senses 2-methylisocitrate levels to regulate bacterial virulence in Pseudomonas aeruginosa.
Sci Adv, 8, 2022
|
|
8JO0
 
 | |
8JNS
 
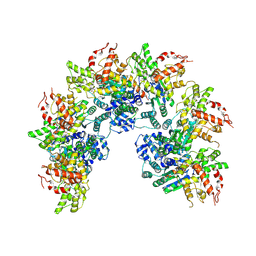 | | cryo-EM structure of a CED-4 hexamer | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | 著者 | Li, Y, Shi, Y. | | 登録日 | 2023-06-06 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural insights into CED-3 activation.
Life Sci Alliance, 6, 2023
|
|
7XP0
 
 | |
8IMX
 
 | | Cryo-EM structure of GPI-T with a chimeric GPI-anchored protein | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xu, Y, Li, T, Qu, Q, Li, D. | | 登録日 | 2023-03-07 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
8IMY
 
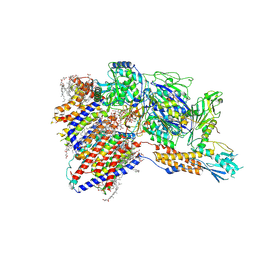 | | Cryo-EM structure of GPI-T (inactive mutant) with GPI and proULBP2, a proprotein substrate | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Li, T, Xu, Y, Qu, Q, Li, D. | | 登録日 | 2023-03-07 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
7JIT
 
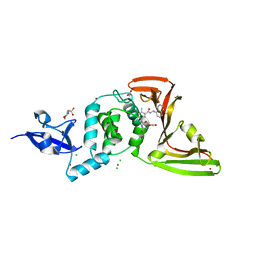 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder495 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(carbamoylcarbamoyl)amino]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIR
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder457 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ACETATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIW
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder530 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIV
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder530 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | 著者 | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | 登録日 | 2022-11-01 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (6.03 Å) | | 主引用文献 | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
