7X37
 
 | | Cryo-EM structure of Coxsackievirus B1 A particle in complex with nAb 2E6 (CVB1-A:2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3E
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 9A3 (CVB1-pre-A:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3D
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 9A3 (CVB1-M:9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X40
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-01 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4K
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | 分子名称: | 9A3 heavy chain, 9A3 light chain, Genome polyprotein, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X46
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X47
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | 分子名称: | 2E6 heavy chain, 2E6 light chain, Genome polyprotein, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3C
 
 | | Cryo-EM structure of Coxsackievirus B1 muture virion in complex with nAbs 8A10 and 5F5 (CVB1-M:8A10:5F5) | | 分子名称: | 5F5 heavy chain, 5F5 light chain, 8A10 heavy chain, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4M
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10, 2E6 and 9A3) | | 分子名称: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | 著者 | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | 登録日 | 2022-03-02 | | 公開日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | 分子名称: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | 著者 | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | 登録日 | 2016-12-19 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
1QCZ
 
 | |
7XGY
 
 | | cryo-EM structure of hemoglobin | | 分子名称: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Liu, N, Wang, H.W. | | 登録日 | 2022-04-07 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Nat.Methods, 20, 2023
|
|
7XY7
 
 | | Adenosine receptor bound to a non-selective agonist in complex with a G protein obtained by cryo-EM | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | 著者 | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | 登録日 | 2022-05-31 | | 公開日 | 2023-05-03 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
7XY6
 
 | | Adenosine receptor bound to an agonist in complex with G protein obtained by cryo-EM | | 分子名称: | 2-[6-azanyl-3,5-dicyano-4-[4-(cyclopropylmethoxy)phenyl]pyridin-2-yl]sulfanylethanamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhang, J.Y, Chen, Y, Hua, T, Song, G.J. | | 登録日 | 2022-05-31 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Cryo-EM structure of the human adenosine A 2B receptor-G s signaling complex.
Sci Adv, 8, 2022
|
|
2I9B
 
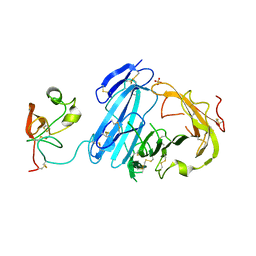 | | Crystal structure of ATF-urokinase receptor complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Urokinase plasminogen activator surface receptor, ... | | 著者 | Lubkowski, J, Barinka, C. | | 登録日 | 2006-09-05 | | 公開日 | 2007-01-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of interaction between urokinase-type plasminogen activator and its receptor.
J.Mol.Biol., 363, 2006
|
|
3BT1
 
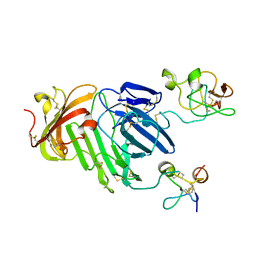 | | Structure of urokinase receptor, urokinase and vitronectin complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, Urokinase-type plasminogen activator, ... | | 著者 | Huang, M. | | 登録日 | 2007-12-27 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes
Nat.Struct.Mol.Biol., 15, 2008
|
|
6HLS
 
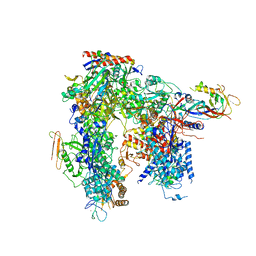 | | Yeast apo RNA polymerase I* | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | 登録日 | 2018-09-11 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HLR
 
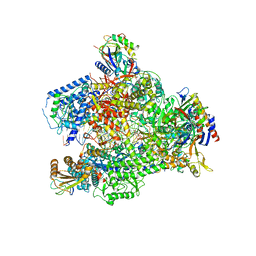 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP (core focused) | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | 登録日 | 2018-09-11 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
3BT2
 
 | | Structure of urokinase receptor, urokinase and vitronectin complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, ... | | 著者 | Huang, M. | | 登録日 | 2007-12-27 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes
Nat.Struct.Mol.Biol., 15, 2008
|
|
7VV9
 
 | |
7VVG
 
 | |
7VVB
 
 | |
7VV8
 
 | |
6HLQ
 
 | | Yeast RNA polymerase I* elongation complex bound to nucleotide analog GMPCPP | | 分子名称: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | 著者 | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | 登録日 | 2018-09-11 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
7WPQ
 
 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 3.27 angstron resolution (2 units) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | 著者 | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | 登録日 | 2022-01-24 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.267 Å) | | 主引用文献 | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
