3X08
 
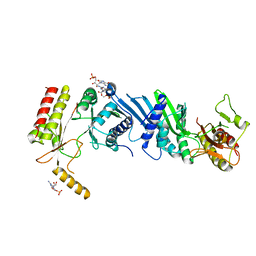 | | Crystal structure of PIP4KIIBETA N203A complex with GMP | | 分子名称: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X07
 
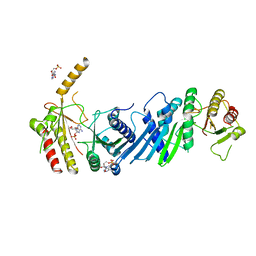 | | Crystal structure of PIP4KIIBETA N203A complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-09 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3WZZ
 
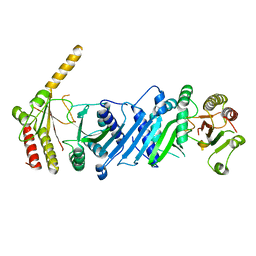 | | Crystal structure of PIP4KIIBETA | | 分子名称: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | 著者 | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | 登録日 | 2014-10-08 | | 公開日 | 2015-10-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
2FBP
 
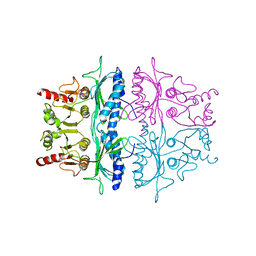 | | STRUCTURE REFINEMENT OF FRUCTOSE-1,6-BISPHOSPHATASE AND ITS FRUCTOSE 2,6-BISPHOSPHATE COMPLEX AT 2.8 ANGSTROMS RESOLUTION | | 分子名称: | FRUCTOSE 1,6-BISPHOSPHATASE | | 著者 | Ke, H, Thorpe, C.M, Seaton, B.A, Marcus, F, Lipscomb, W.N. | | 登録日 | 1990-06-07 | | 公開日 | 1992-04-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution.
J.Mol.Biol., 212, 1990
|
|
6TYF
 
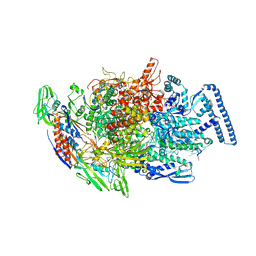 | |
8TB0
 
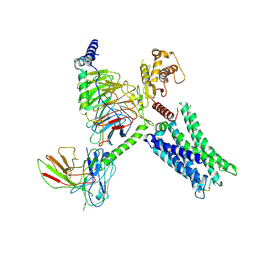 | | Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16 | | 分子名称: | GPR61 fused to dominant negative G alpha S/I N18 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Lees, J.A, Dias, J.M, Han, S. | | 登録日 | 2023-06-28 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
8TB7
 
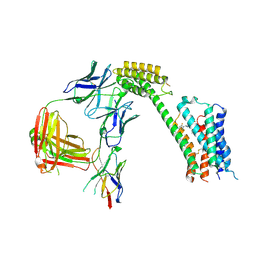 | | Cryo-EM Structure of GPR61- | | 分子名称: | 6-{[(3,5-difluoropyridin-4-yl)methyl]amino}-N-(4-ethoxy-6-methylpyrimidin-2-yl)-2-methoxy-N-(2-methoxyethyl)pyridine-3-sulfonamide, Fab hinge-binding nanobody, Fab24 BAK5 heavy chain, ... | | 著者 | Lees, J.A, Dias, J.M, Han, S. | | 登録日 | 2023-06-28 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
6TYG
 
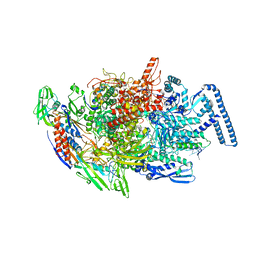 | |
6U3I
 
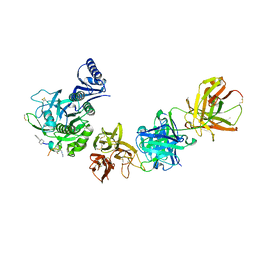 | |
5I5K
 
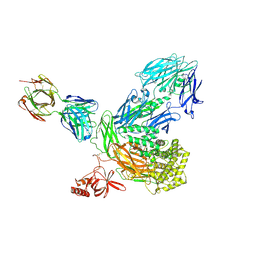 | |
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | 分子名称: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | 著者 | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | 登録日 | 2018-11-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.393 Å) | | 主引用文献 | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6ITQ
 
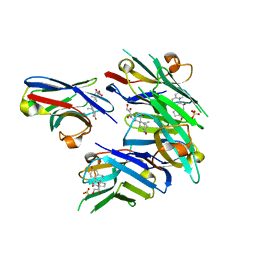 | | Crystal structure of cortisol complexed with its nanobody at pH 10.5 | | 分子名称: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, SULFATE ION, anti-cortisol camelid antibody | | 著者 | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | 登録日 | 2018-11-24 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.526 Å) | | 主引用文献 | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
6U2F
 
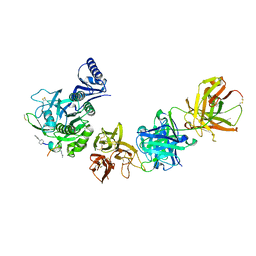 | |
8TTY
 
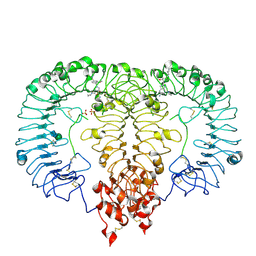 | | Crystal structure of monkey TLR7 ectodomain with compound 5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N~7~-butyl-2-({4-[(cyclobutylamino)methyl]-2-methoxyphenyl}methyl)-2H-pyrazolo[4,3-d]pyrimidine-5,7-diamine, ... | | 著者 | Critton, D.A. | | 登録日 | 2023-08-15 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.101 Å) | | 主引用文献 | Discovery of Novel TLR7 Agonists as Systemic Agent for Combination With aPD1 for Use in Immuno-oncology.
Acs Med.Chem.Lett., 15, 2024
|
|
7CYP
 
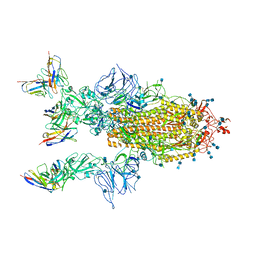 | | Complex of SARS-CoV-2 spike trimer with its neutralizing antibody HB27 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of HB27, ... | | 著者 | Wang, X, Zhu, L. | | 登録日 | 2020-09-04 | | 公開日 | 2021-06-09 | | 最終更新日 | 2022-02-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Double lock of a potent human therapeutic monoclonal antibody against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
8TTZ
 
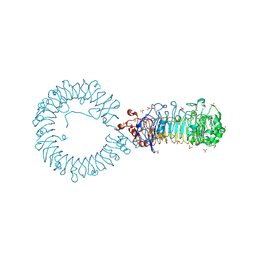 | | Crystal structure of monkey TLR7 ectodomain with compound 20 | | 分子名称: | (3S)-3-({5-amino-1-[(2-methoxy-4-{[(oxan-4-yl)amino]methyl}phenyl)methyl]-1H-pyrazolo[4,3-d]pyrimidin-7-yl}amino)hexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Critton, D.A. | | 登録日 | 2023-08-15 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Discovery of Novel TLR7 Agonists as Systemic Agent for Combination With aPD1 for Use in Immuno-oncology.
Acs Med.Chem.Lett., 15, 2024
|
|
6TYE
 
 | |
8IK2
 
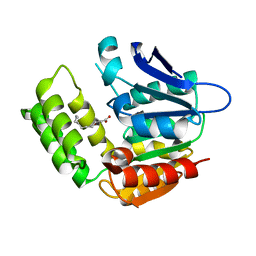 | | RhlA exhibits dual thioesterase and acyltransferase activities during rhamnolipid biosynthesis | | 分子名称: | (3~{S})-3-oxidanyldecanoic acid, 3-(3-hydroxydecanoyloxy)decanoate synthase | | 著者 | Tang, T, Fu, L.H, Xie, W.H, Luo, Y.Z, Zhang, Y.T, Si, T. | | 登録日 | 2023-02-28 | | 公開日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.151 Å) | | 主引用文献 | RhlA Exhibits Dual Thioesterase and Acyltransferase Activities during Rhamnolipid Biosynthesis
Acs Catalysis, 13, 2023
|
|
3F0A
 
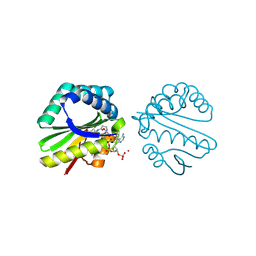 | | Structure of a putative n-acetyltransferase (ta0374) in complex with acetyl-coa from thermoplasma acidophilum | | 分子名称: | ACETYL COENZYME *A, CHLORIDE ION, N-ACETYLTRANSFERASE, ... | | 著者 | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-10-24 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3F9G
 
 | |
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
7DFG
 
 | |
7DFH
 
 | |
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | 著者 | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-16 | | 最終更新日 | 2019-06-05 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
