6MUT
 
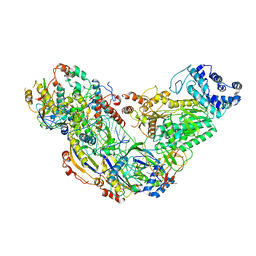 | | Cryo-EM structure of ternary Csm-crRNA-target RNA with anti-tag sequence complex in type III-A CRISPR-Cas system | | 分子名称: | RNA (5'-R(*GP*UP*GP*GP*AP*AP*AP*GP*GP*CP*GP*GP*GP*CP*AP*GP*AP*GP*GP*C)-3'), RNA (5'-R(P*CP*CP*UP*CP*UP*GP*CP*CP*CP*GP*CP*CP*UP*UP*UP*CP*CP*AP*C)-3'), Uncharacterized protein Csm1, ... | | 著者 | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | 登録日 | 2018-10-23 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
7LTO
 
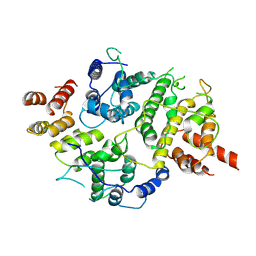 | | Nse5-6 complex | | 分子名称: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | 著者 | Yu, Y, Li, S.B, Zheng, S, Tangy, S, Koyi, C, Wan, B.B, Kung, H.H, Andrej, S, Alex, K, Patel, D.J, Zhao, X.L. | | 登録日 | 2021-02-19 | | 公開日 | 2021-05-19 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Integrative analysis reveals unique structural and functional features of the Smc5/6 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6MJU
 
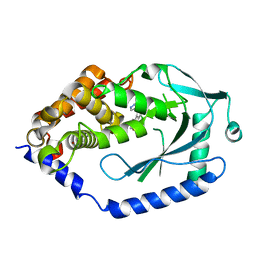 | | human cGAS catalytic domain bound with the inhibitor G108 | | 分子名称: | 1-[6,7-dichloro-9-(1H-pyrazol-4-yl)-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | 著者 | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | 登録日 | 2018-09-22 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
6MJX
 
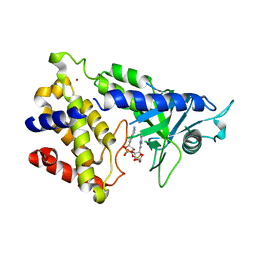 | | human cGAS catalytic domain bound with cGAMP | | 分子名称: | Cyclic GMP-AMP synthase, ZINC ION, cGAMP | | 著者 | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | 登録日 | 2018-09-23 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
3Q0D
 
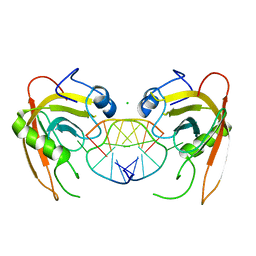 | | Crystal structure of SUVH5 SRA- hemi methylated CG DNA complex | | 分子名称: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*AP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*(5CM)P*GP*TP*CP*AP*G)-3'), ... | | 著者 | Eerappa, R, Simanshu, D.K, Patel, D.J. | | 登録日 | 2010-12-15 | | 公開日 | 2011-02-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3704 Å) | | 主引用文献 | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3PT6
 
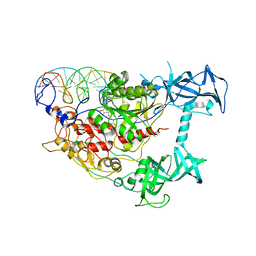 | | Crystal structure of mouse DNMT1(650-1602) in complex with DNA | | 分子名称: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | 著者 | Song, J, Patel, D.J. | | 登録日 | 2010-12-02 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3PTA
 
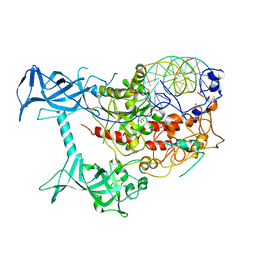 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | 分子名称: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | 著者 | Song, J, Patel, D.J. | | 登録日 | 2010-12-02 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3PT9
 
 | |
1I34
 
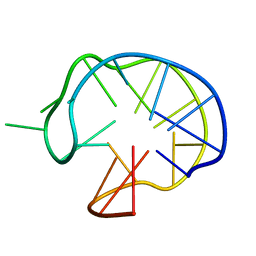 | | SOLUTION DNA QUADRUPLEX WITH DOUBLE CHAIN REVERSAL LOOP AND TWO DIAGONAL LOOPS CONNECTING GGGG TETRADS FLANKED BY G-(T-T) TRIAD AND T-T-T TRIPLE | | 分子名称: | 5'-D(*GP*GP*TP*TP*TP*TP*GP*GP*CP*AP*GP*GP*GP*TP*TP*TP*TP*GP*GP*T)-3' | | 著者 | Kuryavyi, V, Majumdar, A, Shallop, A, Chernichenko, N, Skripkin, E, Jones, R, Patel, D.J. | | 登録日 | 2001-02-12 | | 公開日 | 2001-06-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)-G(syn)-G(anti)-G(anti) tetrads flanked by a G-(T-T) Triad and a T-T-T triple.
J.Mol.Biol., 310, 2001
|
|
3O6E
 
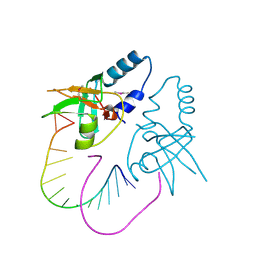 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | 分子名称: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | 著者 | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | 登録日 | 2010-07-28 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.904 Å) | | 主引用文献 | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7MKK
 
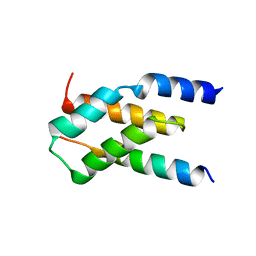 | |
6NXF
 
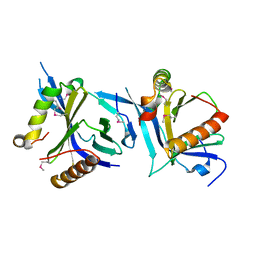 | |
3Q0C
 
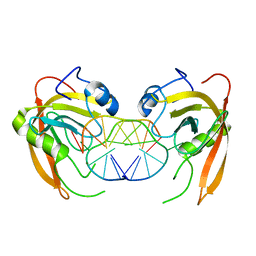 | | Crystal structure of SUVH5 SRA-fully methylated CG DNA complex in space group P6122 | | 分子名称: | DNA (5'-D(*AP*CP*TP*AP*(5CM)P*GP*TP*AP*GP*TP*T)-3'), Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH5, ... | | 著者 | Eerappa, R, Simanshu, D.K, Patel, D.J. | | 登録日 | 2010-12-15 | | 公開日 | 2011-02-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6567 Å) | | 主引用文献 | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
6MUA
 
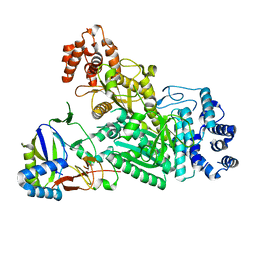 | |
3OWI
 
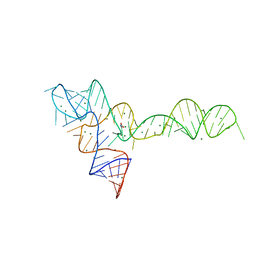 | |
3OXD
 
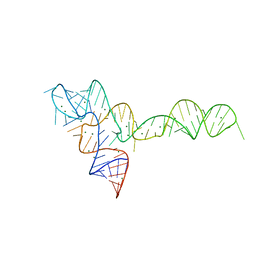 | |
6MRK
 
 | |
6MUU
 
 | | Cryo-EM structure of Csm-crRNA binary complex in type III-A CRISPR-Cas system | | 分子名称: | RNA (25-MER), Uncharacterized protein Csm1, Uncharacterized protein Csm2, ... | | 著者 | Jia, N, Wang, C, Eng, E.T, Patel, D.J. | | 登録日 | 2018-10-23 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
1JVC
 
 | | Dimeric DNA Quadruplex Containing Major Groove-Aligned A.T.A.T and G.C.G.C Tetrads Stabilized by Inter-Subunit Watson-Crick A:T and G:C Pairs | | 分子名称: | 5'-D(*GP*AP*GP*CP*AP*GP*GP*T)-3' | | 著者 | Zhang, N, Gorin, A, Majumdar, A, Kettani, A, Chernichenko, N, Skripkin, E, Patel, D.J. | | 登録日 | 2001-08-29 | | 公開日 | 2001-10-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Dimeric DNA quadruplex containing major groove-aligned A-T-A-T and G-C-G-C tetrads stabilized by inter-subunit Watson-Crick A-T and G-C pairs.
J.Mol.Biol., 312, 2001
|
|
7L9P
 
 | | Structure of human SHLD2-SHLD3-REV7-TRIP13(E253Q) complex | | 分子名称: | Mitotic spindle assembly checkpoint protein MAD2B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pachytene checkpoint protein 2 homolog, ... | | 著者 | Xie, W, Patel, D.J. | | 登録日 | 2021-01-04 | | 公開日 | 2021-03-03 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3OX0
 
 | |
7LHL
 
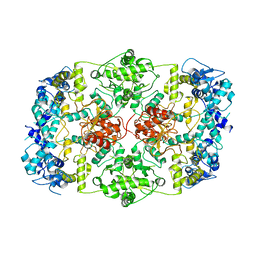 | |
3QZV
 
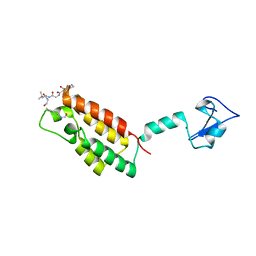 | | Crystal Structure of BPTF PHD-linker-bromo in complex with histone H4K12ac peptide | | 分子名称: | Histone H4, Nucleosome-remodeling factor subunit BPTF, ZINC ION | | 著者 | Li, H, Ruthenburg, A.J, Patel, D.J. | | 登録日 | 2011-03-07 | | 公開日 | 2011-06-01 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3O33
 
 | |
3O34
 
 | |
