8I88
 
 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA complex | | 分子名称: | Piwi domain-containing protein, RNA (5'-R(P*GP*A)-3'), TIR domain-containing protein | | 著者 | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | 登録日 | 2023-02-03 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
1DBX
 
 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase from H. influenzae (HI1434) | | 分子名称: | cysteinyl-tRNA(Pro) deacylase | | 著者 | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 1999-11-03 | | 公開日 | 2000-06-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
9C5Q
 
 | |
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-04 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (4.75 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2025-03-12 | | 実験手法 | ELECTRON MICROSCOPY (3.73 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-04 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.98 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YE5
 
 | | SARS-CoV-2 Spike (6P) in complex with 2 R1-32 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | 登録日 | 2022-07-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (6.75 Å) | | 主引用文献 | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
6Q0R
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | 分子名称: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6Q0W
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to Indisulam and RBM39 | | 分子名称: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OMV
 
 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4-DNA complex | | 分子名称: | AcrVA4, Cpf1, DNA (5'-D(*CP*GP*TP*CP*CP*TP*TP*TP*AP*GP*GP*A)-3'), ... | | 著者 | Chang, L, Li, Z, Zhang, H. | | 登録日 | 2019-04-19 | | 公開日 | 2019-06-12 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6PWC
 
 | | A complex structure of arrestin-2 bound to neurotensin receptor 1 | | 分子名称: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | 著者 | Yin, W, Li, Z, Jin, M, Yin, Y.-L, de Waal, P.W, Pal, K, Gao, X, He, Y, Gao, J, Wang, X, Zhang, Y, Zhou, H, Melcher, K, Jiang, Y, Cong, Y, Zhou, X.E, Yu, X, Xu, H.E. | | 登録日 | 2019-07-22 | | 公開日 | 2019-12-04 | | 最終更新日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | A complex structure of arrestin-2 bound to a G protein-coupled receptor.
Cell Res., 29, 2019
|
|
6Q0V
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to tasisulam and RBM39 | | 分子名称: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
2PY2
 
 | | Structure of Herring Type II Antifreeze Protein | | 分子名称: | Antifreeze protein type II, CALCIUM ION | | 著者 | Liu, Y, Li, Z, Lin, Q, Seetharaman, J, Sivaraman, J, Hew, C.-L. | | 登録日 | 2007-05-15 | | 公開日 | 2007-06-26 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and Evolutionary Origin of Ca-Dependent Herring Type II Antifreeze Protein.
PLoS ONE, 2, 2007
|
|
6OHW
 
 | | Structural basis for human coronavirus attachment to sialic acid receptors. Apo-HCoV-OC43 S | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | 著者 | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | 登録日 | 2019-04-07 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | 分子名称: | Inner capsid protein VP2, RNA-directed RNA polymerase | | 著者 | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | 登録日 | 2019-04-10 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
8VAU
 
 | | Nicotinamide Riboside and CD38: Covalent Inhibition and Live-Cell Labeling | | 分子名称: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Nicotinamide riboside, alpha-D-ribofuranose | | 著者 | Kao, G, Zhang, X.N, Nasertorabi, F, Katz, B.B, Li, Z, Dai, Z, Zhang, Z, Zhang, L, Louie, S.G, Cherezov, V, Zhang, Y. | | 登録日 | 2023-12-11 | | 公開日 | 2024-11-06 | | 最終更新日 | 2024-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Nicotinamide Riboside and CD38: Covalent Inhibition and Live-Cell Labeling.
Jacs Au, 4, 2024
|
|
8W7L
 
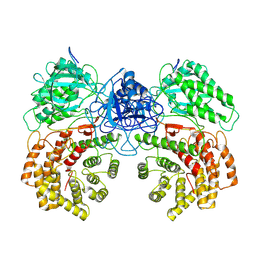 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC mutant H522A. | | 分子名称: | PHOSPHATE ION, PneA, Protein kinase domain-containing protein | | 著者 | Li, Y, Luo, M, Shao, K, Li, J, Li, Z. | | 登録日 | 2023-08-30 | | 公開日 | 2024-09-04 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Structure of PneA bound PneKC at 3.75 Angstroms resolution.
To Be Published
|
|
9IV9
 
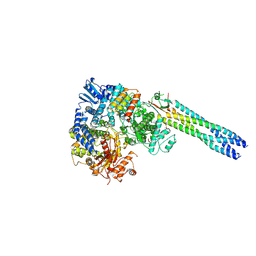 | | Cryo-EM structure of a truncated Nipah Virus L Protein bound by Phosphoprotein Tetramer | | 分子名称: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | 著者 | Xue, L, Chang, T, Gui, J, Li, Z, Zhao, H, Zou, B, Li, M, He, J, Chen, X, Xiong, X. | | 登録日 | 2024-07-23 | | 公開日 | 2025-05-21 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.31 Å) | | 主引用文献 | Cryo-EM structures of Nipah virus polymerase complex reveal highly varied interactions between L and P proteins among paramyxoviruses.
Protein Cell, 2025
|
|
9IVA
 
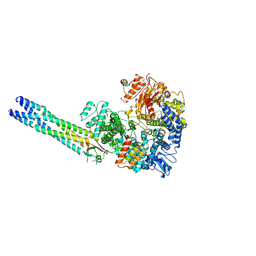 | | Cryo-EM structure of the full-length Nipah Virus L Protein bound by Phosphoprotein Tetramer | | 分子名称: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | 著者 | Xue, L, Chang, T, Gui, J, Li, Z, Zhao, H, Zou, B, Li, M, He, J, Chen, X, Xiong, X. | | 登録日 | 2024-07-23 | | 公開日 | 2025-05-21 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (2.52 Å) | | 主引用文献 | Cryo-EM structures of Nipah virus polymerase complex reveal highly varied interactions between L and P proteins among paramyxoviruses.
Protein Cell, 2025
|
|
1PYM
 
 | |
1T2T
 
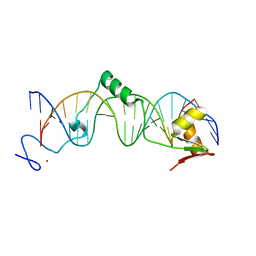 | | Crystal structure of the DNA-binding domain of intron endonuclease I-TevI with operator site | | 分子名称: | 5'-D(*AP*AP*TP*TP*AP*AP*AP*GP*GP*GP*CP*AP*GP*TP*CP*CP*TP*AP*CP*AP*A)-3', 5'-D(*TP*TP*TP*GP*TP*AP*GP*GP*AP*CP*TP*GP*CP*CP*CP*TP*TP*TP*AP*AP*T)-3', Intron-associated endonuclease 1, ... | | 著者 | Edgell, D.R, Derbyshire, V, Van Roey, P, LaBonne, S, Stanger, M.J, Li, Z, Boyd, T.M, Shub, D.A, Belfort, M. | | 登録日 | 2004-04-22 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2N96
 
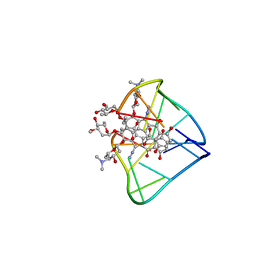 | | An unexpected mode of small molecule DNA binding provides the structural basis for DNA cleavage by the potent antiproliferative agent (-)-lomaiviticin A | | 分子名称: | (1R,1'R,2S,2'S,3R,3'R,5aR,10aR,11a'S)-2'-[(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)oxy]-2,2'-diethyl-11,11 '-dihydrazinyl-6,6',9,9'-tetrahydroxy-4,4',5,5',10,10'-hexaoxo-1,1'-bis{[2,4,6-trideoxy-4-(dimethylamino)-beta-L-arabino -hexopyranosyl]oxy}[2,2',3,3',4,4',5,5',5a,8,10,10',10a,11a'-tetradecahydro-1H,1'H-[3,3'-bibenzo[b]fluorene]]-2-yl 2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranoside, DNA (5'-D(*GP*CP*TP*AP*TP*AP*GP*C)-3') | | 著者 | Woo, C.M, Li, Z, Paulson, E, Herzon, S.B. | | 登録日 | 2015-11-07 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for DNA cleavage by the potent antiproliferative agent (-)-lomaiviticin A.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
