8AWG
 
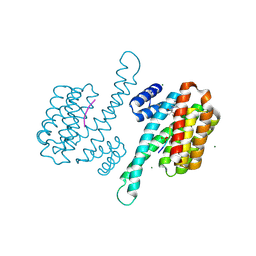 | | small molecule stabilizer for ERalpha and 14-3-3 (1074202) | | 分子名称: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | 著者 | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | 登録日 | 2022-08-29 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AZE
 
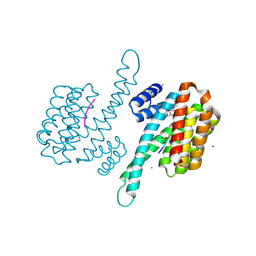 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075306) | | 分子名称: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[1-[(4-chlorophenyl)amino]cyclopentyl]carbonylpiperidin-4-yl]methyl]ethanamide, ERalpha peptide, ... | | 著者 | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | 登録日 | 2022-09-06 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AXE
 
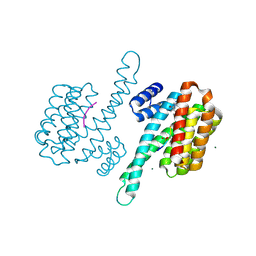 | | Small molecule stabilizer for ERalpha and 14-3-3 (1074210) | | 分子名称: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[2-[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]ethyl]ethanamide, Estrogen receptor, ... | | 著者 | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | 登録日 | 2022-08-31 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
6SKK
 
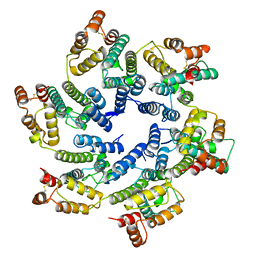 | | Structure of the native full-length HIV-1 capsid protein in helical assembly (-13,8) | | 分子名称: | capsid protein | | 著者 | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | 登録日 | 2019-08-15 | | 公開日 | 2020-08-26 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8EG6
 
 | | huCaspase-6 in complex with inhibitor 2a | | 分子名称: | (3R)-1-(ethanesulfonyl)-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | 登録日 | 2022-09-11 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
8EG5
 
 | | huCaspase-6 in complex with inhibitor 3a | | 分子名称: | (3R,5S)-1-(ethanesulfonyl)-5-phenyl-N-[4-(trifluoromethoxy)phenyl]piperidine-3-carboxamide (bound form), 1,2-ETHANEDIOL, Caspase-6 subunit p11, ... | | 著者 | Zhao, Y, Fan, P, Liu, J, Wang, Y, Van Horn, K, Wang, D, Medina-Cleghorn, D, Lee, P, Bryant, C, Altobelli, C, Jaishankar, P, Ng, R.A, Ambrose, A.J, Tang, Y, Arkin, M.R, Renslo, A.R. | | 登録日 | 2022-09-11 | | 公開日 | 2023-05-10 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Engaging a Non-catalytic Cysteine Residue Drives Potent and Selective Inhibition of Caspase-6.
J.Am.Chem.Soc., 145, 2023
|
|
3H4M
 
 | | AAA ATPase domain of the proteasome- activating nucleotidase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | 著者 | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | 登録日 | 2009-04-20 | | 公開日 | 2009-06-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.106 Å) | | 主引用文献 | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
5UP4
 
 | |
5E8L
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase 11 from Arabidopsis thaliana | | 分子名称: | Heterodimeric geranylgeranyl pyrophosphate synthase large subunit 1, chloroplastic | | 著者 | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | 登録日 | 2015-10-14 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.807 Å) | | 主引用文献 | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
3H43
 
 | | N-terminal domain of the proteasome-activating nucleotidase of Methanocaldococcus jannaschii | | 分子名称: | Proteasome-activating nucleotidase | | 著者 | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | 登録日 | 2009-04-17 | | 公開日 | 2009-06-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
5E8H
 
 | | Crystal structure of geranylfarnesyl pyrophosphate synthases 2 from Arabidopsis thaliana | | 分子名称: | Geranylgeranyl pyrophosphate synthase 3, chloroplastic | | 著者 | Wang, C, Chen, Q, Wang, G, Zhang, P. | | 登録日 | 2015-10-14 | | 公開日 | 2015-11-11 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
5E8K
 
 | | Crystal structure of polyprenyl pyrophosphate synthase 2 from Arabidopsis thaliana | | 分子名称: | Geranylgeranyl pyrophosphate synthase 10, mitochondrial | | 著者 | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | 登録日 | 2015-10-14 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.028 Å) | | 主引用文献 | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
1LQG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE COMPLEX WITH URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | 分子名称: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Saikrishnan, K, Sagar, M.B, Ravishankar, R, Roy, S, Purnapatre, K, Handa, P, Varshney, U, Vijayan, M. | | 登録日 | 2002-05-10 | | 公開日 | 2002-11-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Domain closure and action of uracil DNA glycosylase (UDG): structures of new crystal forms containing the Escherichia coli enzyme and a comparative study of the known structures involving UDG.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | 分子名称: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | 著者 | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | 登録日 | 2015-10-07 | | 公開日 | 2016-03-16 | | 最終更新日 | 2017-08-23 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
6Z18
 
 | | Crystal structure of RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG; R32 form | | 分子名称: | RNA-10mer: CCGG(N4,N4-dimethyl-C)GCCGG | | 著者 | Ruszkowski, M, Sekula, B, Mao, S, Haruehanroengra, P, Sheng, J. | | 登録日 | 2020-05-12 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Base pairing, structural and functional insights into N4-methylcytidine (m4C) and N4,N4-dimethylcytidine (m42C) modified RNA.
Nucleic Acids Res., 48, 2020
|
|
7VM8
 
 | |
3H4P
 
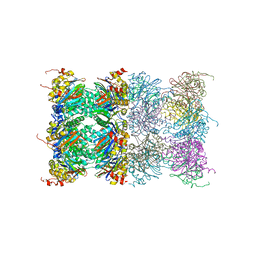 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | 分子名称: | Proteasome subunit alpha, Proteasome subunit beta | | 著者 | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | 登録日 | 2009-04-20 | | 公開日 | 2009-06-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
6QLZ
 
 | | IDOL F3ab subdomain | | 分子名称: | E3 ubiquitin-protein ligase MYLIP | | 著者 | Martinelli, L, Johansson, P, Wan, P.T, Gunnarsson, J, Guo, H, Boyd, H. | | 登録日 | 2019-02-01 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.343 Å) | | 主引用文献 | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
5X41
 
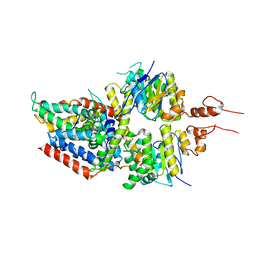 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | 分子名称: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | 著者 | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | 登録日 | 2017-02-09 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.47 Å) | | 主引用文献 | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
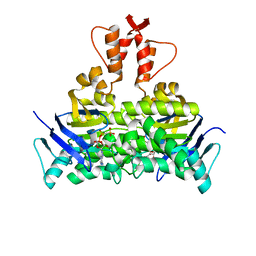 | | Structure of a CbiO dimer bound with AMPPCP | | 分子名称: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | 著者 | Bao, Z, Qi, X, Wang, J, Zhang, P. | | 登録日 | 2017-02-09 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5XN6
 
 | | Heterodimer crystal structure of geranylgeranyl diphosphate synthases 1 with GGPPS Recruiting Protein(OsGRP) from Oryza sativa | | 分子名称: | Os02g0668100 protein, Os07g0580900 protein | | 著者 | Wang, C, Zhou, F, Lu, S, Zhang, P. | | 登録日 | 2017-05-18 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.598 Å) | | 主引用文献 | A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5X3X
 
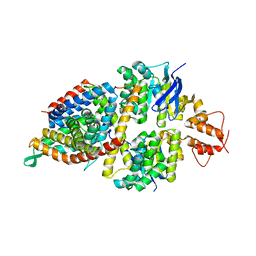 | | 2.8A resolution structure of a cobalt energy-coupling factor transporter-CbiMQO | | 分子名称: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | 著者 | Bao, Z, Qi, X, Wang, J, Zhang, P. | | 登録日 | 2017-02-09 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.788 Å) | | 主引用文献 | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5XN5
 
 | |
4Z7F
 
 | | Crystal structure of FolT bound with folic acid | | 分子名称: | FOLIC ACID, Folate ECF transporter | | 著者 | Zhao, Q, Wang, C.C, Wang, C.Y, Zhang, P. | | 登録日 | 2015-04-07 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.194 Å) | | 主引用文献 | Structures of FolT in substrate-bound and substrate-released conformations reveal a gating mechanism for ECF transporters
Nat Commun, 6, 2015
|
|
5UOT
 
 | |
