5GW4
 
 | | Structure of Yeast NPP-TRiC | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GW5
 
 | | Structure of TRiC-AMP-PNP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
1Z34
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z36
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z33
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase | | Descriptor: | purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z39
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2'-deoxyinosine | | Descriptor: | 2'-DEOXYINOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z38
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with inosine | | Descriptor: | INOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z37
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with adenosine | | Descriptor: | ADENOSINE, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z35
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
9UDE
 
 | | Crystal structure of MonCI in complex with diepoxidized farnesyl acetate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xiao, H.L, Guan, Y.Z, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
9UDB
 
 | | Crystal structure of MonCI in complex with farnesyl acetate | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl acetate, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Deng, Y.M, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
9UDD
 
 | | Crystal structure of MonCI in complex with monoepoxidized farnesyl acetate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xiao, H.L, Guan, Y.Z, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
6GFE
 
 | | High-resolution Structure of a therapeutic full-length anti-NPRA Antibody with exceptional Conformational Diversity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Hoerer, S. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Therapeutic Full-Length Anti-NPRA IgG4 Antibody: Dissecting Conformational Diversity.
Biophys.J., 116, 2019
|
|
8GTW
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound JZD-26 | | Descriptor: | (2S)-4-(3,4-dichlorophenyl)-1-[(2-oxidanylidene-1H-quinolin-4-yl)carbonyl]-N-[3,3,3-tris(fluoranyl)propyl]piperazine-2-carboxamide, 3C-like proteinase | | Authors: | Su, H.X, Nie, T.Q, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of novel inhibitors against SARS-CoV-2 3CLpro
To Be Published
|
|
8GTV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound JZD-07 | | Descriptor: | 3C-like proteinase, 4-[(2~{S})-4-(3,4-dichlorophenyl)-2-(morpholin-4-ylmethyl)piperazin-1-yl]carbonyl-1~{H}-quinolin-2-one | | Authors: | Su, H.X, Nie, T.Q, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of novel inhibitors against SARS-CoV-2 3CLpro
To Be Published
|
|
9IWV
 
 | | Crystal structure of Lsd18 after incubation with the substrate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Wang, Q, Kim, C.Y, Chen, X. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
6KS6
 
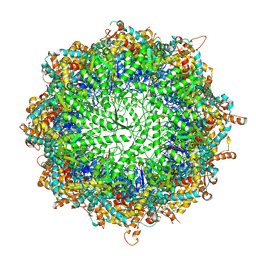 | | TRiC at 0.2 mM ADP-AlFx, Conformation 1, 0.2-C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2019-08-23 | | Release date: | 2019-09-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | An ensemble of cryo-EM structures of TRiC reveal its conformational landscape and subunit specificity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8HM1
 
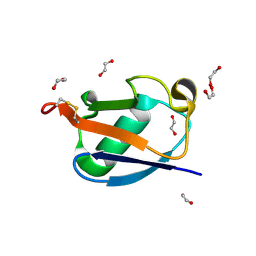 | |
8HM3
 
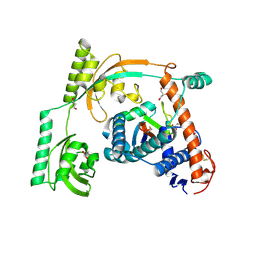 | | Complex of PPIase-BfUbb | | Descriptor: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8HM2
 
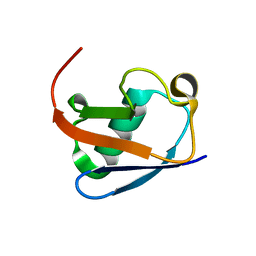 | |
8HM4
 
 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
4N9F
 
 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
3SR9
 
 | | Crystal structure of mouse PTPsigma | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Wang, J, Hou, L, Li, J, Ding, J. | | Deposit date: | 2011-07-07 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the homology and differences between mouse protein tyrosine phosphatase-sigma and human protein tyrosine phosphatase-sigma
Acta Biochim.Biophys.Sin., 43, 2011
|
|
9CS3
 
 | | TRiC-ADP-S2 state is a conformation when TRiC incubated in 1 mM ADP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The conformational landscape of TRiC ring-opening and its underlying stepwise mechanism revealed by cryo-EM.
Qrb Discov, 6, 2025
|
|
9CSA
 
 | | TRiC-ATP-AlFx | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Jin, M, Cong, Y. | | Deposit date: | 2024-07-23 | | Release date: | 2025-01-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The conformational landscape of TRiC ring-opening and its underlying stepwise mechanism revealed by cryo-EM.
Qrb Discov, 6, 2025
|
|
