1UCU
 
 | |
5GKN
 
 | |
3J7T
 
 | | Calcium atpase structure with two bound calcium ions determined by electron crystallography of thin 3D crystals | | Descriptor: | CALCIUM ION, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Yonekura, K, Kato, K, Ogasawara, M, Tomita, M, Toyoshima, C. | | Deposit date: | 2014-08-07 | | Release date: | 2015-02-18 | | Last modified: | 2016-09-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.4 Å) | | Cite: | Electron crystallography of ultrathin 3D protein crystals: atomic model with charges
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6JNT
 
 | | Catalase structure determined by eEFD (dataset 1) | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yonekura, K, Maki-Yonekura, S. | | Deposit date: | 2019-03-18 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | A new cryo-EM system for electron 3D crystallography by eEFD.
J.Struct.Biol., 206, 2019
|
|
6JNU
 
 | | Catalase structure determined by eEFD (dataset 2) | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yonekura, K, Maki-Yonekura, S. | | Deposit date: | 2019-03-18 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.001 Å) | | Cite: | A new cryo-EM system for electron 3D crystallography by eEFD.
J.Struct.Biol., 206, 2019
|
|
3VU3
 
 | |
5ZFU
 
 | | Structure of the ExbB/ExbD hexameric complex (ExbB6ExbD3TM) | | Descriptor: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | Authors: | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
5ZFV
 
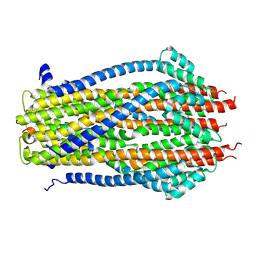 | | Structure of the ExbB/ExbD pentameric complex (ExbB5ExbD1TM) | | Descriptor: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | Authors: | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | Deposit date: | 2018-03-07 | | Release date: | 2018-05-09 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
8ZRT
 
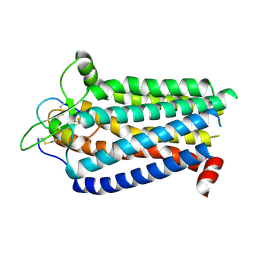 | | Cryo-EM structure focused on the receptor of the ET-1 bound ETBR-DNGI complex | | Descriptor: | Endothelin receptor type B, Endothelin-1 | | Authors: | Tani, K, Maki-Yonekura, S, Kanno, R, Negami, T, Hamaguchi, T, Hall, M, Mizoguchi, A, Humbel, B.M, Terada, T, Yonekura, K, Doi, T. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structure of endothelin ET B receptor-G i complex in a conformation stabilized by unique NPxxL motif.
Commun Biol, 7, 2024
|
|
8XWP
 
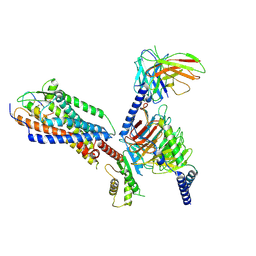 | | Cryo-EM structure of ET-1 bound ETBR-DNGI complex | | Descriptor: | Endothelin receptor type B, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tani, K, Maki-Yonekura, S, Kanno, R, Negami, T, Hamaguchi, T, Hall, M, Mizoguchi, A, Humbel, B.M, Terada, T, Yonekura, K, Doi, T. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure of endothelin ET B receptor-G i complex in a conformation stabilized by unique NPxxL motif.
Commun Biol, 7, 2024
|
|
8XWQ
 
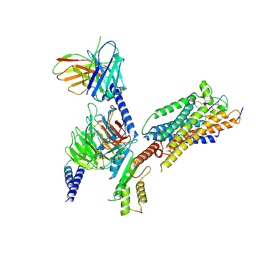 | | Cryo-EM structure of ET-1 bound ETBR-DNGI complex | | Descriptor: | Endothelin receptor type B, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tani, K, Maki-Yonekura, S, Kanno, R, Negami, T, Hamaguchi, T, Hall, M, Mizoguchi, A, Humbel, B.M, Terada, T, Yonekura, K, Doi, T. | | Deposit date: | 2024-01-16 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of endothelin ET B receptor-G i complex in a conformation stabilized by unique NPxxL motif.
Commun Biol, 7, 2024
|
|
8J5A
 
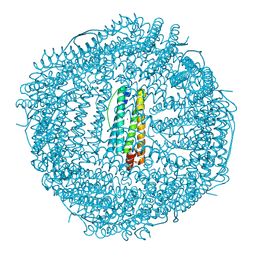 | | Single-particle cryo-EM structure of mouse apoferritin at 1.19 Angstrom resolution (Dataset A) | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Kawakami, K, Maki-Yonekura, S, Hamaguchi, T, Takaba, K, Yonekura, K. | | Deposit date: | 2023-04-21 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (1.19 Å) | | Cite: | Measurement of charges and chemical bonding in a cryo-EM structure.
Commun Chem, 6, 2023
|
|
7VI4
 
 | | Electron crystallographic structure of TIA-1 prion-like domain, A381T mutant | | Descriptor: | TIA-1 prion-like domain | | Authors: | Takaba, K, Maki-Yonekura, S, Sekiyama, N, Imamura, K, Kodama, T, Tochio, H, Yonekura, K. | | Deposit date: | 2021-09-24 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.95 Å) | | Cite: | ALS mutations in the TIA-1 prion-like domain trigger highly condensed pathogenic structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VI5
 
 | | Electron crystallographic structure of TIA-1 prion-like domain, wild type sequence | | Descriptor: | TIA-1 prion-like domain | | Authors: | Takaba, K, Maki-Yonekura, S, Sekiyama, N, Imamura, K, Kodama, T, Tochio, H, Yonekura, K. | | Deposit date: | 2021-09-24 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.761 Å) | | Cite: | ALS mutations in the TIA-1 prion-like domain trigger highly condensed pathogenic structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5ZFP
 
 | |
3A5X
 
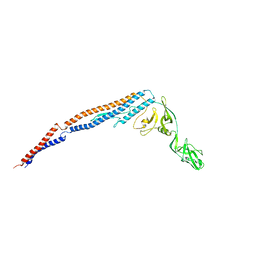 | |
8WEY
 
 | | PSI-LHCI of the red alga Cyanidium caldarium RK-1 (NIES-2137) | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{R} )-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Kato, K, Hamaguchi, T, Nakajima, Y, Kawakami, K, Yonekura, K, Shen, J.R, Nagao, R. | | Deposit date: | 2023-09-19 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | The structure of PSI-LHCI from Cyanidium caldarium provides evolutionary insights into conservation and diversity of red-lineage LHCs.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7Y3F
 
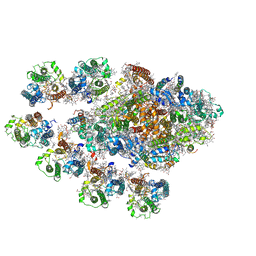 | | Structure of the Anabaena PSI-monomer-IsiA supercomplex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Nagao, R, Kato, K, Hamaguchi, T, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2022-06-10 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of a monomeric photosystem I core associated with iron-stress-induced-A proteins from Anabaena sp. PCC 7120.
Nat Commun, 14, 2023
|
|
7DI8
 
 | |
7CHK
 
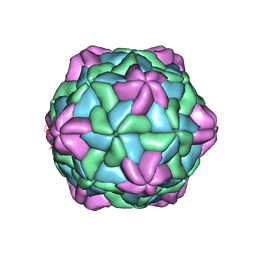 | | Cryo-EM Structure of Apple Latent Spherical Virus (ALSV) | | Descriptor: | VP20 protein, VP24 protein, VP25 protein | | Authors: | Naitow, H, Hamaguchi, T, Maki-Yonekura, S, Isogai, M, Yoshikawa, N, Yonekura, K. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Apple latent spherical virus structure with stable capsid frame supports quasi-stable protrusions expediting genome release.
Commun Biol, 3, 2020
|
|
7VEB
 
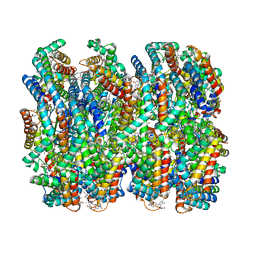 | | Phycocyanin rod structure of cyanobacterial phycobilisome | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Kawakami, K, Hamaguchi, T, Hirose, Y, Kosumi, D, Miyata, M, Kamiya, N, Yonekura, K. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-15 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome.
Nat Commun, 13, 2022
|
|
7VEA
 
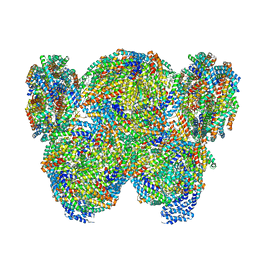 | | Pentacylindrical allophycocyanin core from Thermosynechococcus vulcanus | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Kawakami, K, Hamaguchi, T, Hirose, Y, Kosumi, D, Miyata, M, Kamiya, N, Yonekura, K. | | Deposit date: | 2021-09-08 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Core and rod structures of a thermophilic cyanobacterial light-harvesting phycobilisome.
Nat Commun, 13, 2022
|
|
8HLA
 
 | |
8HH0
 
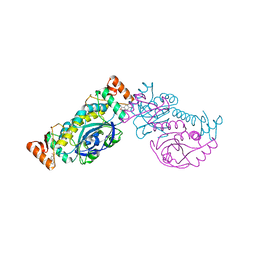 | |
7COY
 
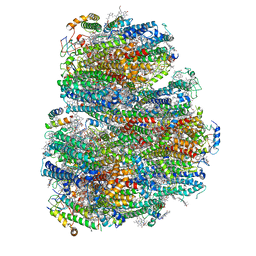 | | Structure of the far-red light utilizing photosystem I of Acaryochloris marina | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL D, ... | | Authors: | Kawakami, K, Yonekura, K, Hamaguchi, T, Kashino, Y, Shinzawa-Itoh, K, Inoue-Kashino, N, Itoh, S, Ifuku, K, Yamashita, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the far-red light utilizing photosystem I of Acaryochloris marina.
Nat Commun, 12, 2021
|
|
